1. Background
The discovery of antibiotics marked the most significant advancement in modern medicine. Antibiotics continue to be the cornerstone of treatment for bacterial infections. The clinical utilization of penicillin G, the first β-lactam antibiotic, spurred the quest for additional derivatives (1). β-lactam antibiotics such as penicillins, carbapenems, and cephalosporins have been extensively employed over recent decades (1). These β-lactam antibiotics inhibit the final step in peptidoglycan synthesis (2). Bacteria have developed an effective mechanism to counteract these antibiotics by hydrolyzing the β-lactam ring using β-lactamase enzymes (3). β-lactamases are categorized into four Ambler classes: Classes A, C, and D (active-site serine β-lactamases) and class B (metallo-β-lactamases or zinc-dependent) (4).
Pseudomonas aeruginosa has emerged as a serious opportunistic pathogen over the past few decades, causing conditions such as cystic fibrosis (CF), urinary tract infections (UTIs), and burn infections (5). Pseudomonas aeruginosa is considered a healthcare-associated infection (HAI) and has been associated with higher fatality rates compared to other bacteria in bloodstream infections (bacteremias) (6, 7). Recent reports have also indicated coinfections of P. aeruginosa in COVID-19 patients (8). Research by Rhoades et al. suggests a higher prevalence of P. aeruginosa in the nasal passages of patients after contracting SARS-CoV-2 (9).
The rise of extensively drug-resistant (XDR), multidrug-resistant (MDR), and pandrug-resistant (PDR) P. aeruginosa strains presents a significant health challenge, leading to increased mortality rates in infections caused by resistant strains (10). Globally, the mortality rate for patients with carbapenem-resistant Pseudomonas infections ranges from 33% to 71% (11). Carbapenem resistance is mediated by various mechanisms, including the production of β-lactamases such as serine β-lactamases (SBLs) and metallo-β-lactamases (MBLs) (12).
Metallo-β-lactamases belong to group 3 of the functional classification (Bush-Jacoby-Medeiros) and class B of the molecular classification (Ambler) (13). Metallo-β-lactamases are enzymes that require zinc (Zn2+) as cofactors for activity and catalyze the hydrolysis of carbapenems (14). The emergence of mobile MBL genes, often carried by class 1 integrons along with other resistance genes, has led to widespread drug resistance (15). Key residues like Trp-87, Phe-61, and Tyr-67 near the second Zn Verona integrin-encoded (VIM)-2 active site create a hydrophobic zone (16). Recent studies have reported interactions between inhibitors and residues such as Arg228, Trp87, and Zn from the VIM-2 active site (17).
The most commonly reported metallo-β-lactamases include VIM, Sao Paulo metalloenzyme (SPM), imipenemase (IMP), German imipenemase (GIM), and New Delhi metallo β-lactamase (NDM) types (18, 19). Currently, VIM is the most prevalent MBL in P. aeruginosa, with VIM-2 being the primary contributor to this predominance (20). The World Health Organization (WHO) has prioritized the discovery of antibiotics or the design of MBL inhibitors to curb the spread of carbapenem-resistant P. aeruginosa strains (21).
Chromosomal mutations are one of the causes of carbapenem resistance in P. aeruginosa (22). Mutations leading to damage in the outer membrane protein OprD create a channel for imipenem penetration, reducing susceptibility to imipenem and other carbapenems (23). Chelating compounds, such as cyclic boronate, ethylenediaminetetraacetic acid (EDTA), sulfamoyl carboxylate, dipicolinic acid derivatives, and thiol group-containing compounds, have demonstrated the ability to inactivate MBLs. However, clinical use inhibitors are yet to be identified (24-26). Overexpression of efflux pump systems like MexAB-OprM, driven by regulatory area mutations, contributes to the mutational derepression of the chromosomal cephalosporinase AmpC, resulting in resistance to meropenem (23).
In recent decades, captopril ((2S)-1-[(2S)-2-Methyl-3-sulfanylpropanoyl] pyrrolidine-2-carboxylic acid; Appendix 1) has been employed for controlling blood pressure. Some previous studies have indicated that captopril can inhibit several MBLs (27, 28). Therefore, captopril can serve as a positive control in our molecular simulations. In 2022, our research team identified ZINC517765 as a promising candidate for inhibiting MBLs (29). Considering the small size and symmetrical structure of ZINC517765, additional efforts have been undertaken to develop and introduce more potent inhibitors.
2. Objectives
This study aims to design a novel potent inhibitor against VIM-2 MBLs of P. aeruginosa using a combination of computer-aided drug design (CADD) and experimental methods. Unlike previous studies, we employed natural compounds and virtual screening alongside molecular dynamics (MD) techniques to discover a potent inhibitor. During the experimental phase, we conducted assays to evaluate and directly compare the inhibitory effects of ZINC04090499 and ZINC517765.
3. Methods
3.1. Molecular Docking
In this study, the 3D structure of VIM-2 MBL (crystal structure, Protein Data Bank (PDB) code: 4C1E, resolution: 1.4 Å) served as the foundation (28). The water molecules within the PDB structure were eliminated using ViewerLite 5.0 software (30). Following the determination of bond ordering, missing hydrogen atoms were inserted. Subsequently, the Gasteiger-Marsili method, a component of the AutoDock Tools package, was employed to calculate the partial atomic charges (31).
For ligand sources, two extensive natural compound libraries, AnalytiCon Discovery NP and IBScreenNP, were utilized. AnalytiCon Discovery NP and IBScreenNP encompass 5154 and 56,799 natural products, respectively. All docking procedures were executed with AutoDock Vina (31), a freely available software facilitating molecular docking and virtual screening. In our research, we adhered to the default parameters of AutoDock Vina. A grid map encompassing the entire active site was defined with dimensions of 30 30 30 points and a grid-point spacing of 1 for all docking operations.
To generate 2D representations of the molecules, we utilized the web-based proteinsPlus program (32). Based on the scoring function of the docking software, the top two compounds (with the lowest docking energy) were chosen for subsequent MD simulations.
3.2. ADMET Analysis
SwissADME was employed to decipher the absorption, distribution, metabolism, and excretion (ADME) characteristics of all previously selected compounds. SwissADME, a web-based program, predicts ADMET properties and enables the evaluation of physicochemical characteristics, water solubility, lipophilicity, drug-likeness, pharmacokinetics, and medicinal chemistry (33). To compute absorption (ABS) (%), Equation 1 was utilized (34, 35):
3.3. Molecular Dynamics Method
Molecular dynamics is a computational technique primarily used for simulating biological systems, particularly for investigating the reconfigurations of biological compounds and their intermolecular interactions in an aqueous environment (36). We conducted MD simulations for (1) unbound VIM-2 MBL; (2) VIM-2 MBL bound to L-captopril (MBL-captopril); (3) VIM-2 MBL bound to ZINC04090499 (MBL-ZINC04090499); and (4) VIM-2 MBL bound to ZINC98363781 (MBL-ZINC98363781), employing the AMBER99SB force field within the GROMACS simulation package (version 2021) (37). Topology files for VIM-2 MBL and the ligands were generated using the AMBER99SB force field and the ACPYPE-Antechamber Python parser interface, respectively (38). The free VIM-2 MBL or ligated VIM-2 MBL systems were placed in cubic boxes and solvated with TIP3P water molecules. Neutralization was achieved by adding 9 Na, 10 Na, 11 Na, and 11 Na ions to the free MBL, MBL-captopril, MBL-ZINC04090499, and MBL-ZINC98363781 systems, respectively. Additionally, all systems (4 simulation runs) underwent energetic minimization until extreme forces dropped below 1000 kJ mol-1 nm-1 on any atom. To accomplish this, the conjugate gradient algorithm followed the steepest descent integrator.
For the treatment of electrostatic and van der Waals interactions (nonbonded and short-range), cutoffs of 0.9 nm and 1.4 nm, respectively, were applied. Long-range electrostatic interactions were managed using the Particle Mesh Ewald (PME) algorithm (39). A 2-fs time step was used, and the linear constraint solver (LINCS) constrained bond lengths. Equilibration of all systems was achieved with a fixed volume ensemble (NVT) and a fixed pressure ensemble (NPT), each for one nanosecond. Trajectory analysis was conducted using VMD (40).
3.4. Principal Component Analysis and Free Energy Landscape
To scrutinize conformational changes, principal component analysis (PCA) calculations were performed. After removing translational and rotational movements, a covariance matrix was constructed using the trajectory data. The eigenvectors and eigenvalues projections were computed using Gmx anaeig and Gmx covar (41). The PCA analysis segregated the enzyme into two conformational subspaces: A physically non-essential and an essential subspace (42). The first two eigenvectors, EigeV1 and EigeV2, were employed for free energy landscape (FEL) analysis following the PCA analysis of the backbone atoms (43).
3.5. Binding Free Energy Evaluation by Molecular Mechanics-Poisson Boltzmann Surface Area
The binding free energy between the inhibitors and the VIM-2 MBL active site was assessed using the free program molecular mechanics-Poisson Boltzmann surface area (MM-PBSA). Mechanics-Poisson Boltzmann surface area is one of the most effective methods in drug design (43). It calculates the binding free energy using Equation 2:
Here, G ligand and G protein represent the ligand and MBL total free energies in the solvent, respectively, while G complex signifies the MBL-ligand energy (44, 45).
3.6. VIM-2 MBL Activity Assay
The MBL activity assay was conducted using a CECIL CE 9500 double-beam spectrophotometer equipped with a thermostat cell holder. Pseudomonas aeruginosa PS679/00 was employed in the assay experiment. Chemicals were sourced from Sigma–Aldrich, Fluka, and Merck, and ZINC04090499 and ZINC517765 were obtained from Molport. The cells were cultured in Trypticase soy broth with a concentration of 0.5 μg/mL of methicillin as a beta-lactamase inducer, incubated at 37°C with shaking. After harvesting the bacteria from the exponential growth phase by centrifugation for 10 minutes at 5000 × g, followed by washing, the cells were suspended in 0.1 M phosphate buffer. The buffer's pH was adjusted to 7 and contained 0.3 mM sucrose, 10 μM ZnCl2, and 7% glycerol as enzyme protecting agents. Cell lysis was achieved through sonication at a frequency of 20 kHz for 10 minutes at 4°C, followed by removal of cellular debris through centrifugation. The resulting supernatant was utilized to assess MBL activity. For the assay, 10 μL of the prepared MBL solution was mixed with an appropriate amount of 0.1 M phosphate buffer and 50 μL of a 0.5 mM benzyl penicillin solution, resulting in a total volume of 1 mL. The beta-lactamase activity was determined by monitoring the absorbance change at 240 nm at 37°C (46). Captopril was employed as a positive control in the assay, and the initial rate of hydrolysis was calculated as a percentage of the benzyl penicillin hydrolysis. The IC50 value was defined as the concentration of each tested compound that resulted in a 50% reduction in this initial rate of hydrolysis (46).
4. Results and Discussion
4.1. Virtual Screening and ADME Investigations of Natural Compounds
As previously mentioned, we gathered a total of 61,953 compounds from the ZINC database. The molecules with the lowest docking energies are listed in Appendix 2. Theoretical ADME calculations were also conducted. SwissADME was utilized to calculate ADME features for each of the selected compounds, and these are presented in Appendix 3. SwissADME provides various computational filters, including Lipinski, Ghose et al. (47), Veber et al. (48), Egan et al. (49), and Muegge et al. (50) rules. None of the listed natural compounds have a synthetic availability rating exceeding ten, making their synthesis relatively straightforward.
For each of the suggested compounds, the estimated octanol/water partition coefficient (log Po/w), a standard indicator of lipophilicity, was accurately determined. The optimal range for log Po/w falls between -0.4 to +5.6 (51, 52). The permissible range for the topological polar surface area (TPSA) is 20 to 130 Å2, and predicting absorption and brain access relies on TPSA (52). Each chemical in our study falls within an appropriate TPSA range. The calculated ABS values for the designed compounds are within acceptable limits. Notably, the docking energy for ZINC98363781 and ZINC04090499 was -14.1 and -12.7 kcal mol-1, respectively, while the positive control, captopril, exhibited an energy of -10.8 kcal mol-1. Based on the docking energies and ADME features, ZINC98363781 and ZINC04090499 were selected as the most promising molecules.
4.2. Investigation of the Active Site of VIM-2 MBL After Molecular Docking
Appendix 4 displays 2D schematics of the selected hits. In Appendix 4A, a single hydrogen bond (H-bond) is observed between captopril and Zn2+. VIM-2 MBL forms four H-bonds with ZINC98363781, involving residues Val211, Asn210, and Zn2+ (Appendix 4B). Notably, in contrast to captopril, ZINC98363781 engages in two H-bond interactions with Zn2+ ions. Appendix 4C reveals three H-bonds between Zn2+ ions and ZINC04090499. Additionally, ZINC04090499 forms four H-bonds, including two with His179 and two with Arg205. Zn2+ ions play a pivotal role in MBL catalysis, initiating catalysis through their binding to the β-lactam ring. For MBL catalysis, it is well-established that the β-lactam molecule bonds with Zn1 via the carbonyl oxygen and the carboxyl group on the 5- or 6-membered fused ring bonds to Zn2 (53). Consequently, strong inhibitor binding to Zn2+ ions can significantly impede MBL activity. The two designed drugs in our study interact with Zn2+ ions through two or more H-bonds, providing an advantage over the positive control drug, captopril.
4.3. Stability of Studied Systems During MD Simulation
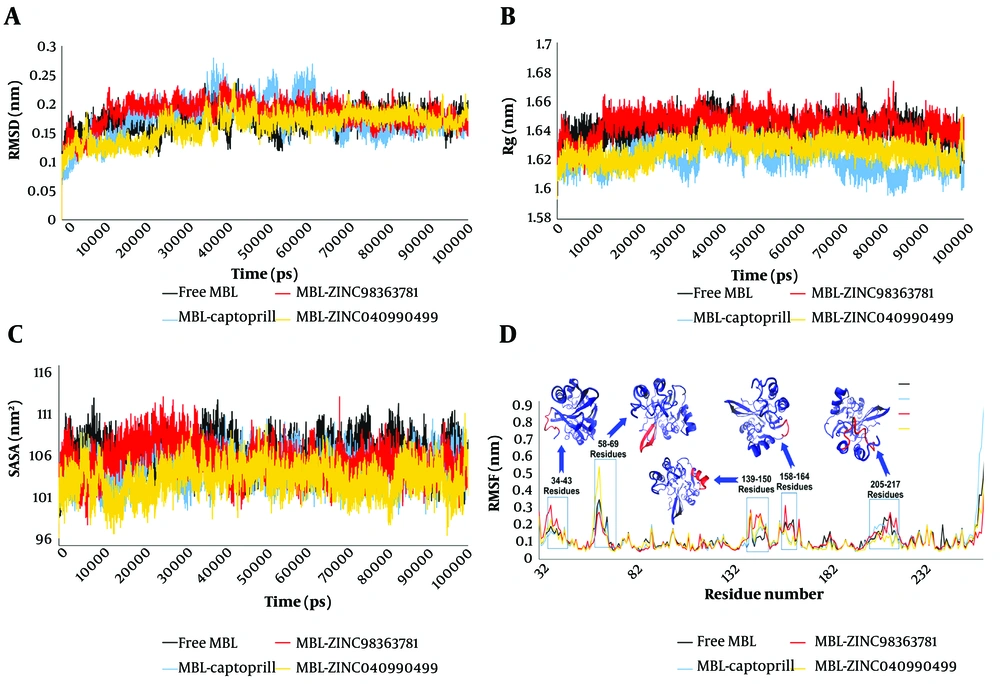
One of the most crucial aspects to evaluate post-MD simulations is the stability of the system. This stability assessment primarily relies on the root mean square deviation (RMSD) value. Figure 1A displays the RMSD values for all the studied systems. As depicted, in the free MBL system, the RMSD value increased to 2.11 Å at 13,940 ps upon the initiation of the simulation. Subsequently, it reached 1.1 Å at 23,880 ps, followed by a temporary surge to 2.36 Å at 38,200 ps. Afterward, a sharp decline occurred, bringing the RMSD value down to 1.21 Å, only to rise again to 2.43 Å at 43,400 ps into the MD simulation. These fluctuations persisted until 70,000 ps, at which point the system attained a relative equilibrium (Figure 1A, black line). In contrast, the MBL-captopril system exhibited higher fluctuations in RMSD values compared to the free MBL system (Figure 1A). It gradually increased from the beginning of the MD simulation until 37,360 ps (2.77 Å), which marked the highest RMSD value in this system. Subsequently, a decrement in the RMSD value was observed, reaching 1.29 Å at 56,050 ps. The next peak in RMSD occurred at 61,590 ps (2.68 Å), followed by a final significant change, resulting in an RMSD value of 1.25 Å at 69,360 ps. Finally, relative equilibrium was established in the MBL-captopril system from 70,000 ps of MD simulation time (Figure 1A, blue line).
The root mean square deviation value (A); calculated Rg values (B); computed solvent-accessible surface area (SASA) values (C); the root mean square fluctuation value (D) for free metallo-β-lactamases (MBL) (black line), MBL-captopril (blue line), MBL-ZINC98363781 (red line), and MBL-ZINC04090499 (yellow line) systems
Root mean square deviation fluctuations in both MBL-ZINC98363781 and MBL-ZINC04090499 complexes were lower compared to the free MBL and MBL-captopril systems (Figure 1A). In the MBL-ZINC98363781 complex, the RMSD score reached 2.28 Å at 11,720 ps, after which no significant shifts were observed, indicating equilibrium (Figure 1A, red line). The MBL-ZINC04090499 system had an RMSD value of 1.86 Å at 6,250 ps. After decreasing to 1.01 Å, it remained relatively stable until 23,780 ps, when it increased to 1.97 Å at 26,880 ps. Subsequently, a few fluctuations led to the highest RMSD value of 2.37 Å at 42,550 ps for MBL-ZINC04090499. Following this, a minor reduction in RMSD occurred, establishing reasonable stability from 50,000 ps until the end of the simulation (Figure 1A, yellow line).
The average RMSD values for free MBL, MBL-captopril, MBL-ZINC98363781, and MBL-ZINC04090499 were 1.69 ± 0.019, 1.73 ± 0.032, 1.83 ± 0.022, and 1.62 ± 0.023 Å, respectively. Consequently, MBL-ZINC04090499 displayed the lowest RMSD value, indicating the highest stability among all systems. While the average RMSD of the MBL-ZINC98363781 complex was higher than that of the MBL-captopril complex, the fewer RMSD fluctuations in the former indicated a more stable equilibrium compared to MBL-captopril.
Another valuable analysis for assessing system stability is the radius of gyration (Rg), which measures the RMSD between the atoms of an intrinsically disordered protein (IDP) and its center of mass (54, 55). Figure 1B illustrates the comparative Rg values for free MBL and ligated MBL systems. The average Rg values were 16.4 Å (free MBL), 16.22 Å (MBL-captopril), 16.43 Å (MBL-ZINC98363781), and 16.27 Å (MBL-ZINC04090499). The average Rg values suggest that the overall structure of VIM-2 MBL remained stable upon binding with captopril and ZINC04090499 molecules. However, the Rg value fluctuations in the MBL-ZINC04090499 complex were less pronounced than those in the MBL-captopril complex. Overall, the Rg values indicate stability across all studied systems. Additionally, the Rg value serves as a feature restricting the energy of the conformational space accessible to the bound molecules.
This perspective is based on the fundamental principle that increasing a protein's buried surface area is necessary for high binding affinity, often achieved by expanding the bound ligand (56). Therefore, a decrease in solvent-accessible surface area (SASA) may correlate with a decrease in Rg value within a system. For further investigation, the computed SASA values for all studied systems are depicted in Figure 1C. The average SASA values were 107.37 nm2 (free MBL), 105.4 nm2 (MBL-captopril), 106.38 nm2 (MBL-ZINC98363781), and 104.16 nm2 (MBL-ZINC04090499). Based on the SASA analysis, the SASA value closely correlates with the Rg value.
Furthermore, a decrease in SASA suggests enhanced protein packing and greater stability (56). Thus, the SASA data indicate that the MBL-ZINC04090499 complex exhibits the highest stability in this study. Appendix 5 displays the crystal structure of MBL-ZINC98363781 and MBL-ZINC04090499 during the MD simulation for further examination.
4.4. Flexibility of Studied Systems During MD Simulation
Residue flexibility was investigated using the root mean square fluctuation (RMSF) for each residue. The P. aeruginosa VIM-2 MBL enzyme (PDB ID: 4C1E) comprises 231 residues, and the RMSF values for all residues are illustrated in Figure 1D. In MD simulations, residues with higher RMSF values exhibit greater flexibility, whereas those with lower values demonstrate reduced motion (57). Each system exhibits five distinct regions in the RMSF plot, each with varying RMSF values.
The first region encompasses residues 34 - 43, where the MBL-ZINC98363781 complex demonstrates the highest RMSF value. Notably, Glu38 exhibits the highest flexibility in the MBL-ZINC98363781 complex with a value of 3.07 Å. The second region encompasses residues 58 - 69, which form the β-hairpin motif. In this region, the MBL-ZINC04090499 system displays the highest RMSF value. Specifically, Asp63 exhibits the highest RMSF value in the MBL-ZINC04090499 complex at 5.27 Å. Interestingly, the RMSF values for Tyr67 in the free MBL and MBL-captopril systems were higher than in the two designed drugs from this study. The next region comprises residues 139 - 150, forming an α-helix.
Ligand binding increases the flexibility of these residues compared to unbound MBL. The fourth region is a loop consisting of residues 158 - 164. Flexibility in this region decreases upon binding of captopril and ZINC04090499 compared to free MBL. However, the binding of ZINC98363781 does not reduce the RMSF value compared to free MBL; in fact, an increase in RMSF is observed in Ser128. Another loop comprises residues 205 - 217, often playing a crucial role in defining protein structure and ligand binding (58). This loop contains some active site residues (Arg205, Asn210, Ala212, and Asp213).
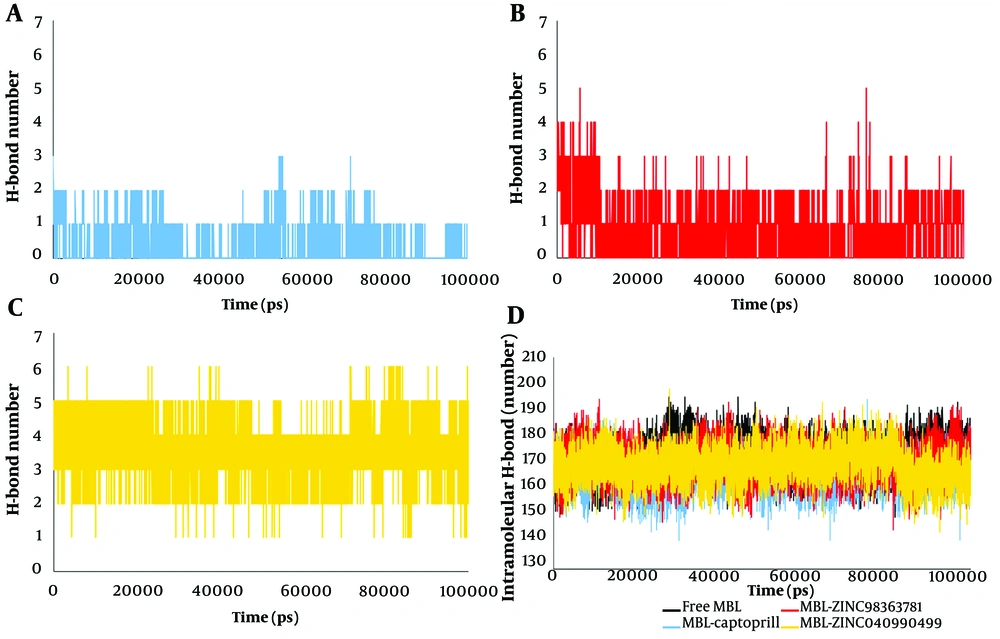
In this loop, the RMSF value shows the greatest reduction after ZINC04090499 binding compared to ZINC98363781 and captopril. This decrease in flexibility in the active site residues can be attributed to the stronger binding of ZINC04090499 to the MBL active site. The number of hydrogen bonds formed by ZINC04090499 within the active site cavity is higher than the other two ligands (Figure 2). The hydrogen bond count further supports the stronger binding of the ZINC04090499 compound to the MBL active site, resulting in reduced RMSF values for active site residues.
4.5. Intramolecular and Intermolecular Hydrogen Bond Calculation
Proteins typically form numerous intramolecular hydrogen bonds between residues, with an average of 1.1 intramolecular hydrogen bonds per residue. Previous studies have shown that intramolecular hydrogen bonds contribute to stabilizing protein structures (59, 60). The intramolecular and intermolecular hydrogen bonds are depicted in Figure 2. The average number of intramolecular hydrogen bonds was 172.33, 166.39, 170.8, and 169.65 for the free MBL, MBL-captopril, MBL-ZINC98363781, and MBL-ZINC04090499 systems, respectively. Consequently, more intramolecular hydrogen bonds were observed in the MBL-ZINC98363781 and MBL-ZINC04090499 systems compared to the MBL-captopril complex (Figure 2D). The presence of additional intramolecular hydrogen bonds suggests enhanced stability in the complexes.
Intermolecular hydrogen bonds formed between the protein and ligands were calculated as intermolecular hydrogen bonds. These hydrogen bonds are crucial for various protein functions, including enzyme catalysis, protein-ligand binding strength, and protein folding (61-63). Figure 2A, B, and C reveal that the number of hydrogen bonds between our designed drugs (ZINC04090499 and ZINC98363781) and VIM-2 MBL active site residues is significantly higher than with captopril. Additionally, the number of hydrogen bonds for ZINC04090499 exceeds that of ZINC98363781. The increased quantity of hydrogen bonds indicates greater affinity between ZINC04090499 and ZINC98363781 for VIM-2 MBL compared to captopril.
4.6. PCA
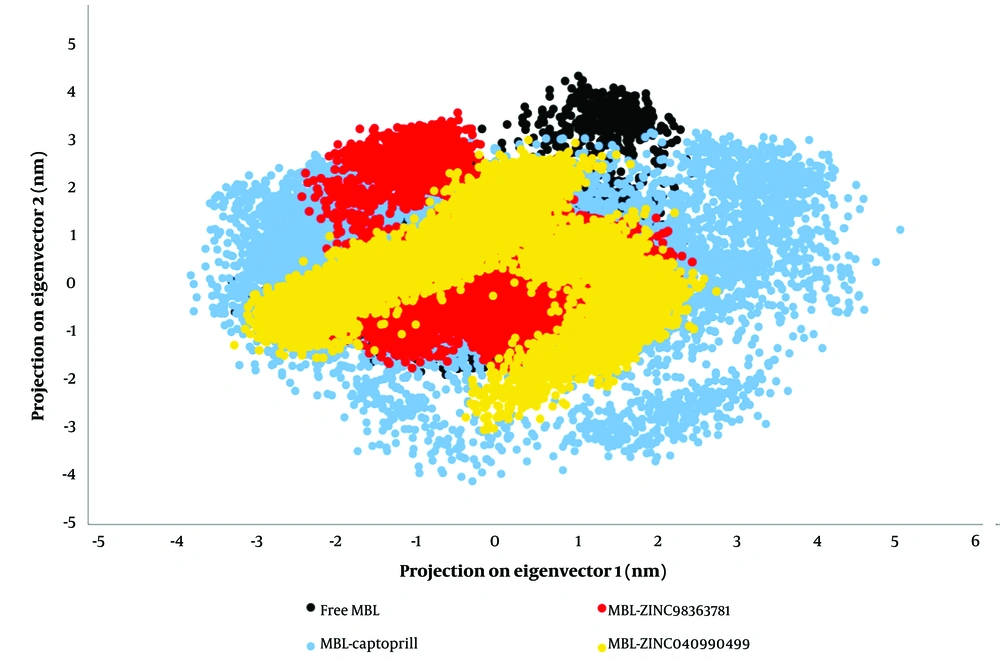
Enzymes derive their specific functions from collective atomic movements, making the evaluation of enzyme stability essential. Principal component analysis was employed to identify motion patterns in the studied systems. By computing the eigenvectors from an MD simulation covariance matrix, it is possible to determine the dominant motions during an MD simulation. Often, a few low-frequency eigenvectors can capture a significant portion of the overall macromolecule fluctuation with large eigenvalues (64). In Figure 3, the Cα atom is projected along eigenvectors 1 and 2 to visualize the conformational sampling of unbound and bound MBL in the relevant subspace. The results indicate that MBL binding with ZINC98363781 and ZINC04090499 exhibits different conformational fluctuations compared to captopril binding (Figure 3).
The occupied conformational space of the MBL-ZINC98363781 and MBL-ZINC04090499 complexes is reduced, indicating greater stability than the MBL-captopril complex. The abrupt change in the occupied conformational space of the MBL-captopril complex may explain the observed high RMSD for MBL-captopril (Figure 1A, blue line).
4.7. FEL
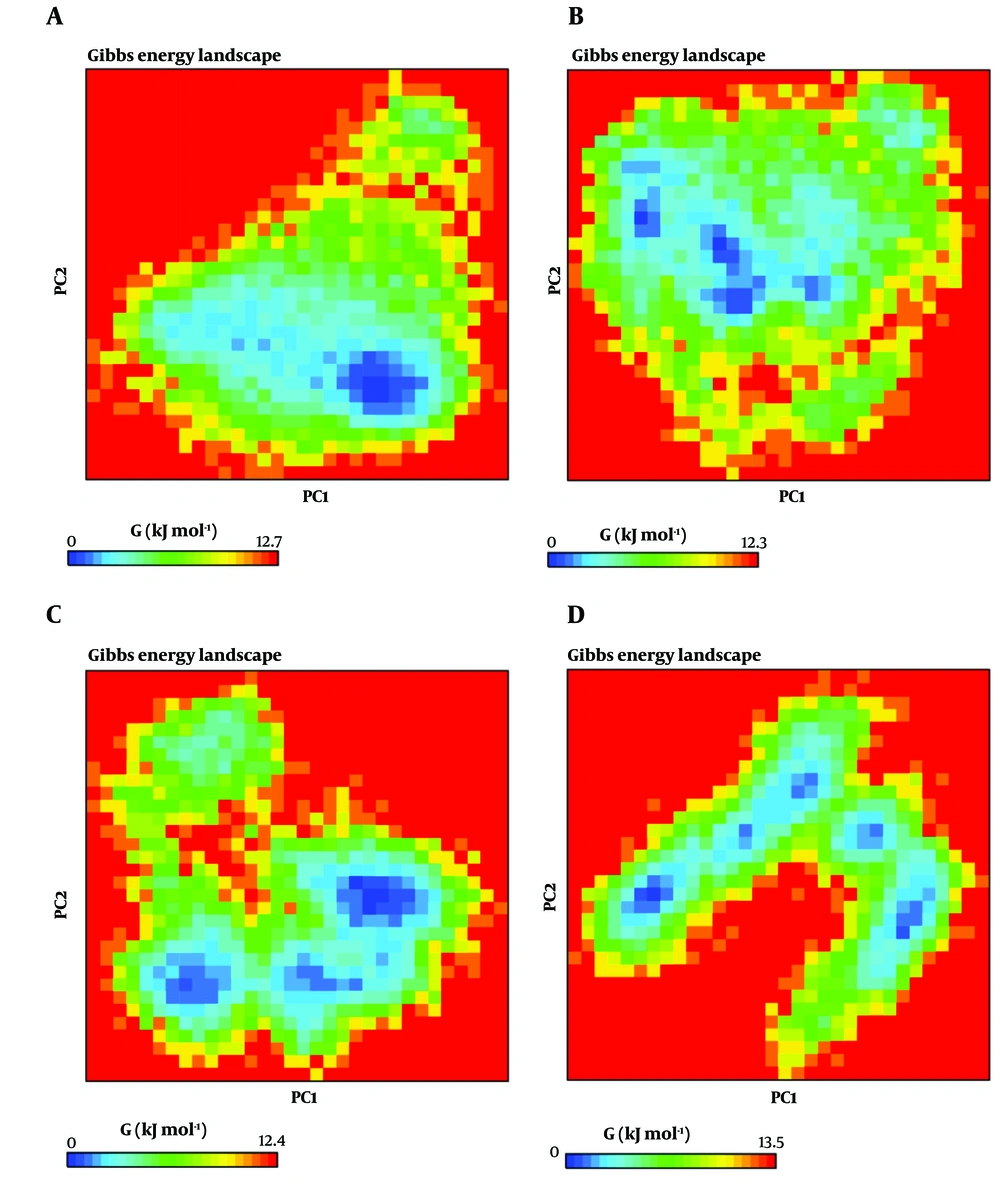
The FEL plot for PC1 and PC2, generated by gmx_anaeig, is presented in Figure 4. For free MBL, MBL-captopril, MBL-ZINC98363781, and MBL-ZINC04090499, the Gibbs energy values vary from 0 to 12.7, 0 to 12.3, 0 to 12.4, and 0 to 13.5, respectively. A shallow and narrow energy basin indicates low system stability (60, 65). In the case of MBL-ZINC98363781 and MBL-ZINC04090499, three distinct deep and broad valleys are observed, while the MBL and MBL-captopril systems display a cluster of three energy basins close to each other. MBL-ZINC98363781 exhibits energy levels similar to the MBL-captopril complex, suggesting that these complexes undergo energetically favorable transitions between structures. The binding of ZINC98363781 and captopril to VIM-2 MBL leads to an increase in the global minima (low-energy basins) of VIM-2 MBL during MD simulations, indicating that these systems are thermodynamically more favorable than free MBL and the MBL-ZINC04090499 complexes.
Free energy landscape (FEL) calculation of free metallo-β-lactamases (MBL) (A); MBL-captopril (B); MBL-ZINC98363781 (C); and MBL-ZINC04090499 (D) systems. The deeper blue spots indicate the principal components where the energy is minimum, and the red spots indicate the principal components where the energy is maximum.
4.8. Free Binding Energy Computed by MM-PBSA Method
Following the simulations, MM-PBSA was utilized to calculate the binding free energy of the compounds (Table 1). The results indicate that the binding free energies of captopril, ZINC98363781, and ZINC04090499 are, respectively, -29.39 ± 5.92 kcal mol-1, -79.74 ± 67.51 kcal mol-1, and -99.65 ± 26.52 kcal mol-1 (Table 1). Consequently, the MBL-ZINC04090499 complex exhibits the lowest binding free energy. Additionally, only the MBL-ZINC04090499 complex has a solvent-accessible surface area energy greater than the MBL-captopril complex. In comparison to the MBL-captopril complex, the MBL-ZINC04090499 complex displays lower electrostatic energy. Furthermore, ZINC98363781 also exhibits lower free binding energy than captopril.
| Energy (kcal mol-1) | MBL-Captopril | MBL-ZINC98363781 | MBL-ZINC04090499 |
|---|---|---|---|
| ∆Evdwa | -155.39 ± 4.81 | -77.62 ± 48.81 | -199.58 ± 53.09 |
| ∆Eelectb | -64.77 ± 7.35 | -58.08 ± 44.18 | -68.06 ± 18.46 |
| ∆Esolvc | 210.69 ± 15.17 | 64.05 ± 57.68 | 180.17 ± 52.46 |
| ∆ESASAd | -19.92 ± 0.27 | -8.09 ± 5.10 | -12.18 ± 5.72 |
| ∆Gbinding | -29.39 ± 5.92 | -79.74 ± 67.51 | -99.65 ± 26.52 |
a Solvent-accessible surface area energy.
b Polar solvation energy.
c Electrostatic energy.
d Van der Waal energy.
4.9. VIM-2 MBL Inhibition by ZINC04090499
To assess and compare the inhibitory potency of ZINC04090499 with our previously identified compound, ZINC517765 (29), assays were conducted to determine the IC50 values of both compounds. Experimental assays revealed that ZINC04090499 has an IC50 value of 25 μM, indicating a stronger inhibitory effect on VIM-2 MBL compared to ZINC517765, which has an IC50 value exceeding 100 μM. This significant difference underscores ZINC04090499's enhanced efficiency in reducing the enzyme's activity by 50%, emphasizing its potential as a potent therapeutic agent. ZINC04090499 contains an indole group linked to butyroyl aspartic acid. The indole moiety's stable, planar structure facilitates strong π-π interactions and hydrogen bonding. The aspartic acid component adds a carboxylic acid functional group, forming strong ionic interactions and hydrogen bonds crucial for binding to metal ions in metalloenzymes like MBLs. In contrast, ZINC517765 consists of two furan rings connected by a methylene bridge, with each furan ring bearing a carboxylic acid group. Furan rings are smaller and less planar than indole, potentially leading to weaker π-π interactions.
4.10. Conclusions
The development of BLs inhibitors represents a potent strategy to safeguard β-lactam antibiotics from BLs. Recently, reports have indicated an increased presence of P. aeruginosa in the bodies of COVID-19 patients. Furthermore, it has been demonstrated that ciprofloxacin-resistant P. aeruginosa in lung abscesses of COVID-19 patients complicates their treatment (66). In this context, we propose a potent inhibitor targeting P. aeruginosa VIM-2 MBL through a combination of computational and experimental studies.
In our study, two compounds, namely ZINC98363781 and ZINC04090499, were selected based on molecular docking studies, and further investigations were conducted on both molecules. Ultimately, we recommend ZINC04090499, a natural substance, as a VIM-2 MBL inhibitor based on the results of MD simulations. The docking energy of ZINC04090499 when binding to the VIM-2 MBL active site is lower (-12.7 kcal mol-1) compared to the positive control (-10.8 kcal mol-1). Furthermore, in comparison to the captopril complex, the suggested inhibitor complex exhibits a reduced RMSD value.
Enzyme assays were conducted to compare the inhibitory effects of ZINC04090499 and ZINC517765. The experimental results align with the predictions of our computational simulations. ZINC04090499, with its larger and more complex structure featuring an indole ring and aspartic acid, may confer stronger and more specific interactions with MBLs, thereby leading to more effective inhibition compared to ZINC517765, which is smaller and based on a furan structure.
According to the MD data, the presence of ZINC04090499 in the VIM-2 MBL active cavity results in a reduction in Rg. This decrease may be attributed to the enhancement of intramolecular H-bonds within the protein following ZINC04090499 binding. In contrast to the unbound MBL and the MBL-captopril complex, the RMSF decreases following ZINC04090499 binding.
Moreover, the number of H-bonds formed between VIM-2 MBL and ZINC04090499 is higher than that with captopril, indicating a strong binding of ZINC04090499 to the active site. The atomic motions were evaluated using the PCA method. Based on the PCA data, the MBL-ZINC04090499 system exhibits reasonable stability compared to the MBL-captopril system. In conclusion, the binding free energy determined by MM-PBSA highlights ZINC04090499's potential to inhibit VIM-2 MBL activity. Nevertheless, further in vitro and in vivo tests are required to evaluate its suggested therapeutic utility fully.