1. Background
Klebsiella pneumoniae, as a frequently reported gram-negative pathogen, causes different types of infections, such as pneumonia, intra-abdominal infection, and bloodstream infection (1, 2). Since the first case of carbapenemase-producing K. pneumoniae infection was described in the United States in 2001 (3), sporadic cases and the outbreak of carbapenem-resistant K. pneumoniae (CRKP) have been reported worldwide (4, 5). The mechanism of antimicrobial resistance (AMR) to carbapenems includes enzyme production, efflux pumps, and porin mutations, all of which cause carbapenem resistance (6, 7), while the production of carbapenemase enzymes is the major mechanism in CRKP (8). In China, the reported isolation rate of CRKP was 19.6% in 2019 (9).
In previous studies, the same sequence type (ST) K. pneumoniae strains were isolated from multiple patients (10-12). Similarly, among all clinical K. pneumoniae isolates from specific wards during a 1-year period, 8 - 20% of STs were isolated from > 1 patient (13, 14). It is not surprising that multidrug-resistant (MDR) K. pneumoniae is more likely to cause nosocomial infections than susceptible strains (14, 15). Furthermore, MDR K. pneumoniae, such as CRKP, leads to significant adverse public health due to less effective antibiotics. The production of class A β-lactamases (such as K. pneumoniae carbapenemase [KPC]), which are capable of hydrolyzing carbapenems, is the major carbapenem resistance mechanism of CRKP, while blaKPC-2 is the main carbapenemase gene in China (16). Whole genome sequencing (WGS) is an effective tool to investigate the epidemiology of pathogen infection by affording a comprehensive population genetic structure of pathogen and strain relatedness (14, 17). Furthermore, K. pneumoniae occupies distinct niches, including host-associated and non-host–associated environments, and exhibits phenotypic and genetic divergence (18).
There is considerable variability in the prevalence of CRKP among countries, regions, and even among different medical institutions (19, 20). In a previous study, the hospital level was found to be associated with the prevalence of CRKP (9). As the majority of the Chinese population is served by non-tertiary hospitals, in-depth characterization of the clinical and genomic epidemiology of CRKP in non-tertiary hospitals can help inform future AMR control programs. Furthermore, few studies have investigated the spread of CRKP isolates in non-tertiary hospitals in mainland China.
2. Objectives
We combined clinical and whole genomic data to analyze the clinical outcomes, antibiotic resistance profiles, and virulent factors of CRKP infection during a 1-year period. Another aim of this study was to find the disseminated clones of CPKP in our hospital and nearby hospitals.
3. Methods
3.1. Study Population, Disease Definitions, and Data Collection
Patients hospitalized at Huzhou Central Hospital between 1 January 2021 and 31 December 2021 were eligible for this cohort study. The inclusion criterion was patients above 18 years who were diagnosed with an infection caused by CRKP (including bacteremia, pneumonia, wound infection, abdominal infection, and so on). The diagnoses of these infections were based on the diagnostic criteria of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) (21). Clinical data were obtained from the hospital’s electronic medical record system, while the microbiology labs in the region provided the antimicrobial susceptibility results. The clinical data of each patient included 3 broad components: (1) basic information (such as age, sex, and comorbidity), (2) antibiotic prescriptions within 30 days of infection with K. pneumoniae, and (3) hospitalization and patient outcomes.
3.2. Definition
Nosocomially acquired infection was defined as infectious diseases acquired 48 hours after hospitalization (22). CRKP was defined as K. pneumoniae that was resistant to ertapenem, meropenem, and/or imipenem (23). To distinguish between CRKP infection and colonization, patients diagnosed with pneumoniae should have a temperature above 38.5°C, white blood cell (WBC) count ≥ 12000 WBC/mm3 or < 4000 WBC/mm3, and one of the following 3 features: (1) new purulent sputum or changes in sputum properties and (2) K. pneumoniae isolated from sputum (3) without other recognized cause.
3.3. Bacterial Isolates
Clinical non-duplicated K. pneumoniae isolates were collected from our hospital from 1 January to 31 December 2021; some isolates were randomly collected from other 2 hospitals (Hospital B and Hospital C) in the same region during the same time period. All K. pneumoniae strains were identified by VITEK 2 GN cards (bioMérieux SA, Marcy-l’Etoile, France) and confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS; Bruker Microflex LT, Bruker Daltonik GmbH, Bremen, Germany). The isolates exhibiting resistance to one of the carbapenems were retained for further research.
3.4. Antibiotic Susceptibility Test
The minimal inhibitory concentrations (MICs) of 16 antibiotics were determined by the agar dilution method or broth dilution method. Piperacillin, cefoperazone, ceftriaxone, ceftazidime, cefoxitin, moxalactam, aztreonam, amikacin, gentamicin, ciprofloxacin, levofloxacin, tigecycline, trimethoprim/sulfamethoxazole, minocycline, tazobactam, sulbactam, and avibactam were purchased from Dalian Meilun Biotech (Dalian, China). Polymyxin B and glucose-6-phosphate (G-6-P) were obtained from Sigma-Aldrich (St Louis, MO, USA). The results of MICs were interpreted according to the Clinical and Laboratory Standards Institute (CLSI; M100 ED32) criteria or European Committee on Antimicrobial Susceptibility Testing (https://www.eucast.org). The MIC breakpoints of tigecycline were issued by the US Food and Drug Administration (susceptible, ≤ 2µg/mL; resistant, ≥ 8 µg/mL). Escherichia coli ATCC25922, K. pneumoniae ATCC BAA1705, and K. pneumoniae ATCC 27853 were used as quality control.
3.5. DNA Extraction, WGS, and Data Analysis
All CRKP isolates were cultured in Luria-Bertani broth at 37°C overnight; DNA extraction was performed as described previously (24). The WGS of strains was performed using an Illumina HiSeq PE150 platform (Novogene Bioinformatics Technology Co, Ltd, Beijing, China). The whole genome sequences of the isolates were analyzed using Kleborate version 2.1.0 (25), and the STs and K (capsule) serotype prediction were analyzed by the Kaptive option (26). The AMR profile, plasmid replicons, and multilocus ST (MLST) were carried out using ResFinder version 4.1 in the Center for Genomic Epidemiology database (27). The virulence genes were screened using the Virulence Factor Database (http://www.mgc.ac.cn/VFs/). The sequencing data for CRKP isolates were deposited in GenBank under accession number PRJNA876055.
3.6. Transmission Analysis
For transmission analysis, we considered that isolates with 0 to 10 single-nucleotide polymorphisms (SNPs) were genetically linked. Patients were considered to have ward contact who shared time in the same ward (CRKP isolated within 4 weeks). Patients were considered to have possible ward contamination who were admitted to the same ward up to 4 weeks apart. Patients were considered to have hospital contact in the same hospital at the same time but not in the same ward.
3.7. Statistical Analysis
Antimicrobial susceptibility data were analyzed using the WHONET version 5.6 (WHO, Geneva, Switzerland). Among patient characteristics, categorical variables were expressed as proportions, while continuous variables were expressed as mean ± SD or median and interquartile range (IQR).
4. Results
4.1. Clinical Features of Patients with CRKP Infection
Forty-nine patients aged > 18 years with a positive carbapenem-resistant K. pneumoniae culture and a clinically established infection were enrolled in the study; these included 33 patients from Huzhou Central Hospital, 13 from Hospital B, and 3 from Hospital C in the same city. The median age of patients was 66.0 years, and 85.7% were male (Table 1). More than 75% of patients had hospital-acquired pneumonia caused by the organism (Table 1). CRKP isolates were obtained from patients in Huzhou Central Hospital, mostly from the intensive care unit (ICU; 39.4%, 13/33). Three-fourths of patients were hospitalized for more than 2 weeks. All patients received antibiotic treatment; 77.6% (38/49) received antibiotic monotherapy, while 10.2% (5/49) received a combination of more than 3 classes of antibiotics. β-Lactam and β-lactamase inhibitor combinations (BLBLICs) were the most common therapy (55.1%, 27/49), while 14.3% (7/49) of the patients were treated with carbapenems. The 30-day mortality was 24.5% (12/49).
| Variables | Values |
|---|---|
| Age, y | 66.0 ± 16.4 |
| ≤ 50 | 9 (18.4) |
| 51 – 60 | 9 (18.4) |
| 61 – 70 | 8 (16.3) |
| 71 – 80 | 15 (30.6) |
| ≥ 81 | 8 (16.3) |
| Sex | |
| Male | 42 (85.7) |
| Female | 7 (14.3) |
| Baseline comorbidities | |
| Heart disease | 3 (6.1) |
| Cerebral vascular accident | 6 (12.2) |
| Chronic kidney disease | 1 (2.0) |
| Dementia | 1 (2.0) |
| COPD | 16 (32.7) |
| Liver disease | 5 (10.2) |
| Diabetes mellitus | 11 (22.5) |
| Non-hematological malignancy | 1 (2.0) |
| Hematological malignancy | 1 (2.0) |
| Hypertension | 14 (28.6) |
| Infection caused by the organism | |
| Lower respiratory tract infection | 37 (75.5) |
| Bloodstream infection | 5 (10.2) |
| Urinary tract infection | 3 (6.1) |
| Surgical wound infection | 4 (8.2) |
| Antibiotics application before CRKP infection | |
| Second-generation cephalosporins | 4 (8.2) |
| Third-generation cephalosporins | 7 (14.3) |
| BLBLICs | 27 (55.1) |
| Carbapenems | 7 (14.3) |
| Fluoroquinolones | 9 (18.4) |
| Amikacin | 1 (2.0) |
| Polymyxin B | 1 (2.0) |
| Cefoxitin | 1 (2.0) |
| Fluconazole | 4 (8.2) |
| Using more than 3 antibiotic classes | 5 (10.2) |
| Clinical outcomes | |
| Cure | 37 (75.5) |
| Death | 12 (24.5) |
| Hospital stay | |
| < 14 | 12 (24.5) |
| ≥ 14 | 37 (75.5) |
Abbreviations: COPD, chronic obstructive pulmonary disease; CRKP, carbapenem-resistant Klebsiella pneumoniae; BLBLICs, β-lactam, and β-lactamase inhibitor combinations.
a Values are expressed as mean ± SD or No. (%).
4.2. Antimicrobial Resistance Phenotypes and Determinants of CRKP
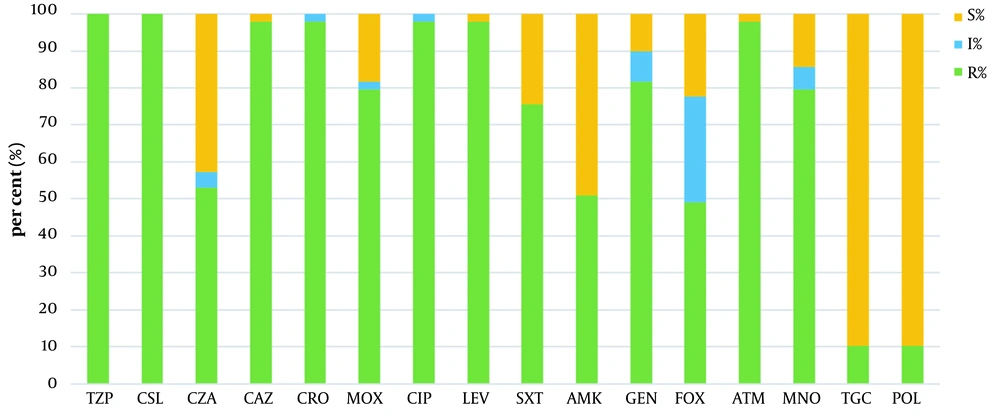
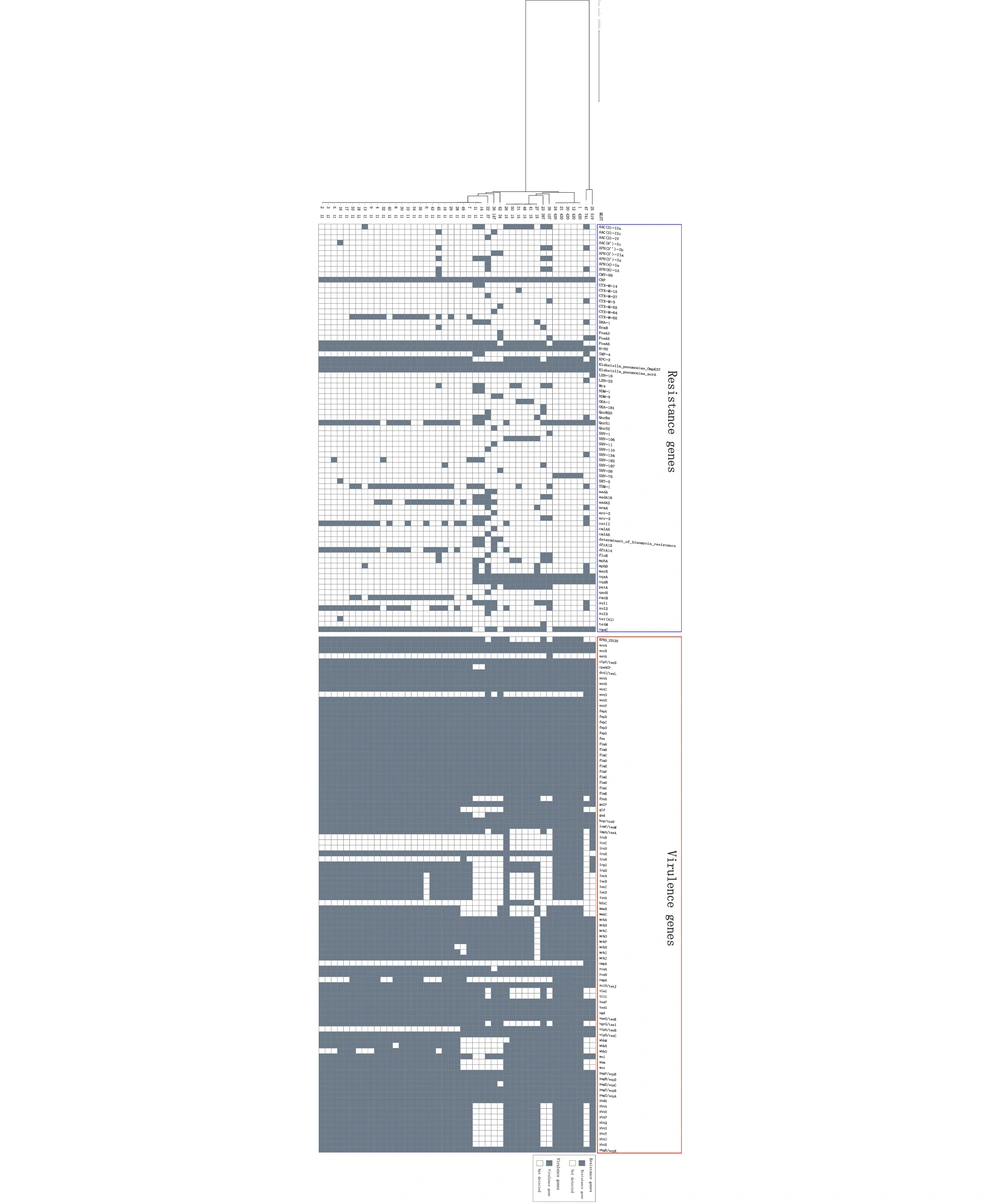
Antimicrobial susceptibility rates of 49 CRKP isolates are shown in Figure 1. Twenty-one (42.8%) out of 49 CRKP isolates were susceptible to ceftazidime-avibactam (CAZ/AVI) in vitro, while 89.8% (44/49) of strains were susceptible to both tigecycline and polymyxin B. According to the 45 strains that were successfully sequenced, the predominant carbapenem-resistant gene was blaKPC-2 (n = 42; 93.3%), followed by blaIMP-4 (n = 3; 6.6%), blaNDM-1 (n = 2; 4.4%), blaNDM-5 (n = 2; 4.4%), and blaOXA-181 (n = 1; 2.2%). Two isolates co-producing IMP-4 and NDM-1 were discovered. The distribution patterns of resistance genes in these strains are shown in Figure 2. We identified the blaCTX−M−65 group as common β-lactamase genes. Furthermore, 100% resistance genes were found in fosfomycin (n = 45), followed by 97.8% in fluoroquinolone (n = 44), 75.6% in aminoglycoside (n = 34), 68.9% in sulfonamide (n = 31), and 64.4% in phenicol (n = 29). Antimicrobial resistance genes, such as fosA-like (associated with fosfomycin resistance), qnrS-like, oqxB-like (associated with fluoroquinolone resistance), and aadA2, rmtB-like (associated with aminoglycoside resistance), were predominant across these strains.
4.3. Sequence Types, Capsular Serotypes, Plasmid Profiling, and Virulent Profiles of CRKP Isolates
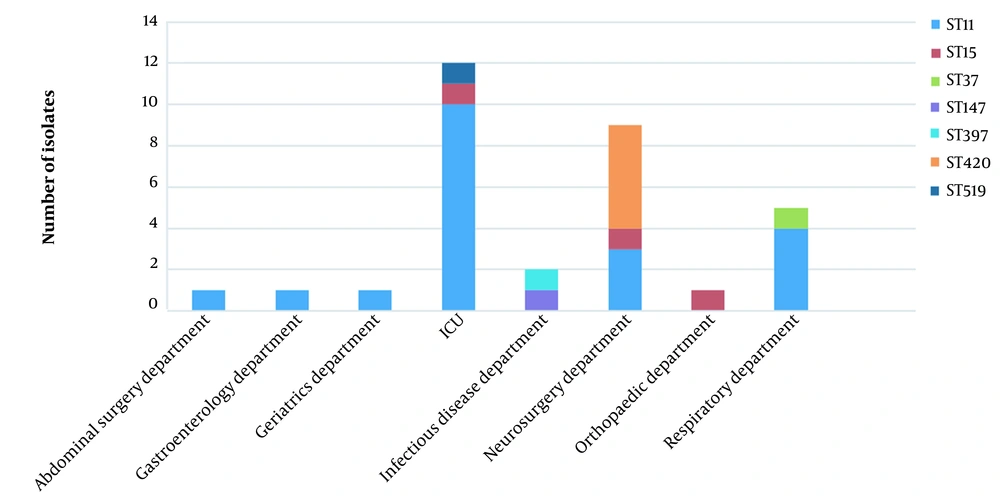
Among the 45 CRKP isolates, 10 different STs were identified. Approximately 60.0% of these belonged to ST11 (n = 27), followed by ST15 (n = 6; 13.3%) and ST420 (n = 5; 11.1%). Other clinical strains were comprised of individual STs, ST34, ST37, ST107, ST147, ST397, ST519, and ST741. ST11 strains were most commonly isolated from ICU, while most ST420 strains were isolated from the neurosurgery department (Figure 3). The majority of strains belonged to KL64 (n = 23) in this study; the other strains were identified as KL119 (n = 6), KL20 (n = 5), KL47 (n = 2), KL21 (n = 1), KL24 (n = 1), KL55 (n = 1), KL81 (n = 1), KL102 (n = 1), KL155 (n = 1), KL158 (n = 1), KL183 (n = 1), and 1 isolate with unknown K type. All KL64 isolates belonged to ST11, while all KL20 belonged to ST420.
To understand whether the AMR determinant was transmitted through the plasmid, we then investigated the types of plasmids. In this regard, 21 plasmid replicon types were found in 45 isolates. From the metadata, the estimated number of plasmids per strain ranged from 1 to 7, and 46.7% (n = 20) of isolates harbored at least 6 plasmids. These 21 plasmid replicon types are classified as incompatibility plasmids (Inc) and mobilizable colicin plasmid (Col) groups. In Inc groups, the plasmid IncF group was the most frequent type, similar to previous reports (Appendix 1 in Supplementary File) (28, 29). The ColRNAI plasmid was the most prevalent plasmid of Col-type.
Different virulence factors in each isolate are shown in Figure 2. The 3 most important siderophore systems in K. pneumoniae strains (ie, enterobactin [entB], yersiniabactin [irp1 and irp2], and aerobactin [iucABCD]) were analyzed. The entB gene was observed in all strains, irp1 and irp2 were detected in 91.1% (41/45), and iucABCD genes were detected in 77.8% (35/45). Most isolates of ST11 and all of ST420 were detected by iutA and rmpA, which are considered to be the main virulence factors (30, 31). Co-occurrence of iutA, rmpA, and iroBCDE genes was observed in KL20 serotypes belonging to ST420.
4.4. Phylogenetic Analysis and Genetic Diversity of CRKP Isolates
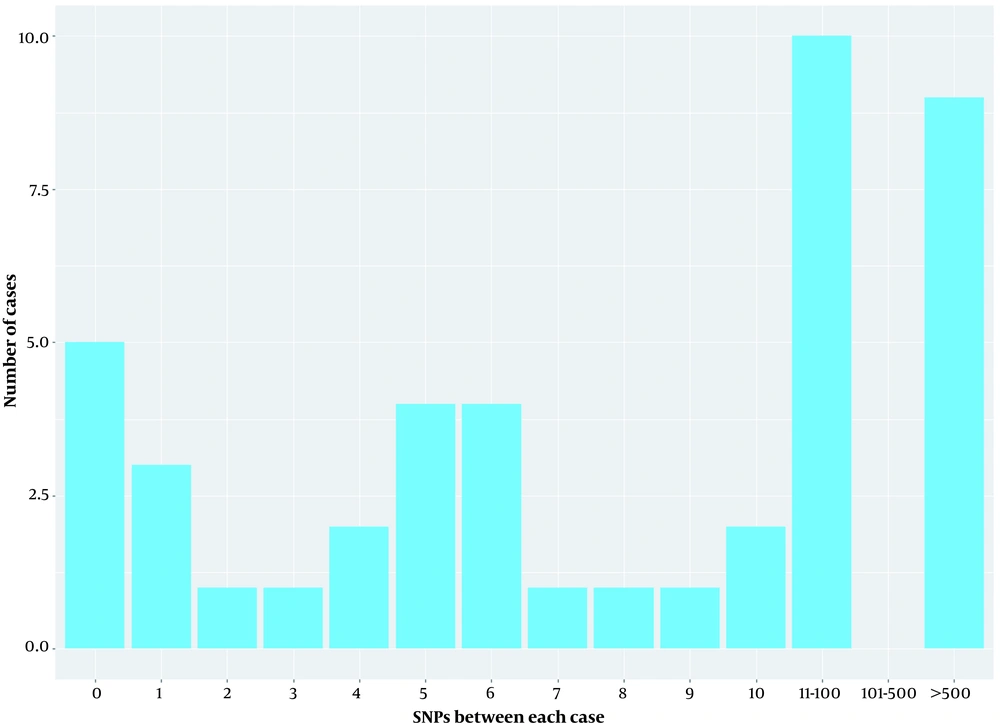
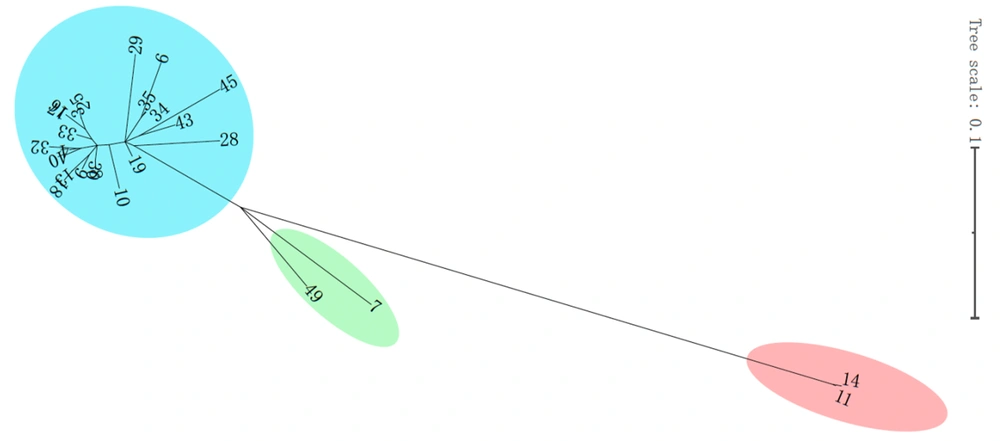
Comparative genomics of microbial genomes helped to characterize the spread of CRKP strains in the setting during the study period; other CRKP strains obtained from the same region were also sequenced. Based on core genome SNPs, a maximum-likelihood phylogenetic tree was reconstructed, and 45 CRKP strains were partitioned into 2 major clades and further delineated into many clusters (Figure 2). Clade 1 was comprised of only 2 strains (ST519 and ST741), while clade 2 was comprised of other strains. Clade 2 was partitioned into 2 subclades; ST420 and ST15 were in subclade 2-1, and ST11 was in subclade 2-2. To elucidate the relationship between the 45 CRKP strains, 10 SNPs were set as the threshold for the genetic linkages between each case. The phylogenetic analysis of CRKP showed that some of these clones were related while others were not. Up to 56.8% (25/44) of case pairs were genetically linked to the genetically related previous case (SNPs ≤10), while 20.5% (9/44) of the isolates were genetically completely unrelated to all previous isolates (SNPs ≥ 100; Figure 4).
4.5. Nosocomial Transmission of CRKP Isolates
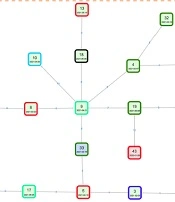
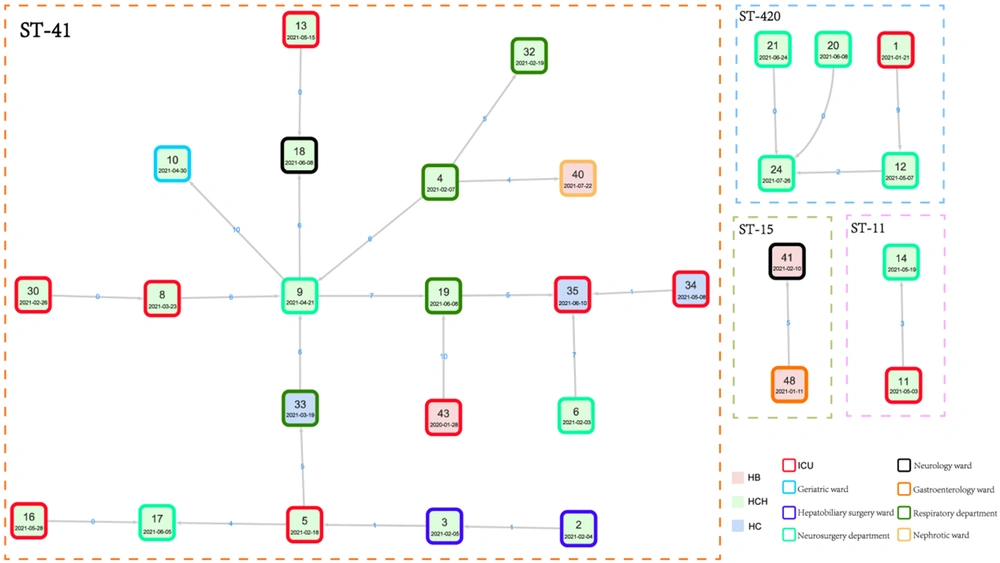
One of the major routes for pathogen transmission is the nosocomial transmission. Strains belonging to the same clade were considered to have a transmission relationship. As ST11 was the most common ST, 27 ST11 isolates were clustered into 3 major subclades, including subclade 1, subclade 2, and subclade 3, with 2, 2, and 23 strains in each subclade, respectively (Figure 5). Based on SNPs ≤ 10, a total of 4 outbreak clusters were classified, of which the largest cluster was comprised of 20 isolates of ST11 (Figure 6). According to the transmission networks, strain 9, isolated from patient 9 (P9) admitted to the neurosurgery department, was the center of the largest cluster. Two sources of this clone were discovered, one from P30 admitted to the ICU, and another from P2 admitted to the abdominal surgery department. Then, this clone was disseminated to other departments in the hospital and even to other hospitals (strains 43 and 35) in the same region. For another 5 patients, ST420 isolates were detected, indicating a transmission from 21 January 2021 to 26 July 2021. For this transmission, the first isolate was discovered in the ICU and then circulated mainly in the neurosurgery department. ST11 isolates were detected in P11 and P14, indicating a transmission chain caused by another ST11 clone. In Hospital B, the same clones of ST15 were isolated from P41 and P48, indicating a small chain of transmission. These results showed that some CRKP clones, especially ST11, were transmitted between different wards in the same hospital and even between different hospitals in the same region.
5. Discussion
The emergence of CRKP has increased worldwide over the last decades (32). In mainland China, the first reported CRKP case dates back to 2004, and it was associated with the K. pneumoniae strain carrying blaKPC-2 (33). Here, we report the molecular epidemiological characteristics of CRKP and identify several outbreaks caused by the organism in a non-tertiary hospital in China during 1-year CRKP surveillance. Despite many advances in infection control measures, nosocomial pneumonia ranks second with an incidence of 5 - 20 cases per 1000 admissions (34). In our study, most patients with CRKP infection had nosocomial pneumonia, which was different from other Chinese CRKP studies focusing on bloodstream infection (35).
It has been reported that mucosal barrier damage can result in CRKP colonizing the respiratory tract and causing infection because of the frequent use of invasive procedures (36). Due to the lack of effective antimicrobial treatment, morbidity and mortality rates have shown a disproportionate increase compared to infections caused by non-carbapenem–resistant K. pneumoniae (37). In this study, the 30-day mortality was 24.5%, which was higher than that in previous reports (38). As ICU admission, mechanical ventilation, and central venous catheterization have been considered independent risk factors for CRKP (39), the high mortality is not surprising as most of the patients in our study were high-risk patients. Another possible explanation is that the patients in our cohort are elderly, which has been found to be associated with 30-day mortality (40).
Previous carbapenem exposure has been identified as an independent risk factor for CRKP (41); however, only a few patients in our cohort were treated with carbapenems before CRKP infection. Intensive care unit patients face a significant risk of CRKP infection because of the ability of CRKP to maintain infectivity over the hospital environment, which makes transmission in the environment and between ICU staff members and patients (42), as CRKP was mainly detected in ICU, neurosurgery, and respiratory departments, which was similar with other reports (43-45). According to these results, a stay in the ICU or surgery wards may be a risk factor for infection with CRKP.
Consistent with previous reports in China (46, 47), most CRKP isolates in this study were KPC-2 producing, and most CRKP isolates were susceptible to tigecycline and polymyxin B, suggesting a therapeutic option (48). However, a previous study conducted in 2018 found a tigecycline resistance rate of 13.1% among CRKP isolates; moreover, treatment failure of polymyxin monotherapy for CRKP infections and colistin-resistant CRKP has recently been reported (49). Ceftazidime-avibactam is a novel BLBLIC with good antibacterial activity against CRKP producing class A and C and some class D carbapenemases, but not metallo–β-lactamases (MBLs) (50). Five of the CAZ/AVI-resistant isolates produced MBL (NDM-1, NDM-5, and IMP-4), of which 3 produced both MBLs and KPC-2. However, the rate of resistance to CAZ/AVI was higher in our study than in other reports (51). Resistance to CAZ/AVI has been linked to specific mutations in the blaKPC gene (52) or blaCTX-M-14 gene (53), while another study has attributed the high expression of blaKPC to low-level CAZ/AVI resistance (54). Our results were in line with this, as the MIC for nearly all CAZ/AVI-resistant strains was below 32 µg/mL.
Plasmids belonging to IncF-type and carrying genes (which encode extended-spectrum β-lactamases [ESBL], carbapenemases, and other genes associated with aminoglycoside and fluoroquinolone) are the most common type. In a previous study, blaKPC-2 genes were frequently found in the IncFII/IncR plasmid replicons (46). Our research also observed a similar phenomenon, as these plasmids were the most frequently discovered. Numerous factors increase K. pneumoniae virulence, such as siderophores (scavenge essential iron), lipopolysaccharide (providing serum resistance), and extracellular polysaccharide capsules (for evasion and inhibition of phagocytosis) (55). While K64 was the dominant K type, no K1 or K2 strains were detected, which are often associated with a highly virulent strain (56). RKP K64, mainly ST11, has been reported to be highly virulent when the strains acquire virulence plasmids (57, 58). We found that the CRKP K64 strain carried rmpA2 and iucA genes in this study. Therefore, acquiring these virulent isolates in our hospital is of clinical concern as dissemination in the ICU is a distinct possibility. Continuous monitoring of these clones may help control their spread.
With the increasing use of carbapenems in clinical settings worldwide, the cloning spread of CRKP in health care settings poses a major clinical concern. ST11 is the dominant ST of CRKP both in Asia and China (46), which has led to a global epidemic since its emergence (59). In this 1-year surveillance study, ST11 was the main epidemic clone, and clonal dissemination was identified mostly in the ICU and neurology departments, which is in line with previous studies (60). CRKP isolates tend to spread rapidly in the neurology wards and ICU due to exposure to more antibiotics and invasive therapy; thus, active surveillance should be taken in these high-risk departments. As CRKP commonly contaminates the environment, such as ventilator- and sink-related sites and bedside tables, the implementation of disinfection should be strengthened (61).
ST11 strains were observed to be transmitted between patients in the same ward, hospital, and region. Two ST11 clones caused 2 transmissions, which is similar to a previous study in which ST11 clones caused an outbreak in a single hospital (62). Our research also showed that ST11 was grouped into 3 subclades, indicating that the ST11 lineage is the clone complex. Furthermore, we also found potential dissemination of the ST420 clone, indicating that ST420 may be another major high-risk clone in our region besides ST11. Several studies have demonstrated that genes, such as rmpA, iucABCD, iutA, and iroBCDN, are associated with hypervirulent K. pneumoniae (63, 64), and all these genes were detected in ST420 isolates of this study.
This study has some limitations. First, we did not perform active surveillance during the study period, and only a small proportion of isolates in the same region were collected. Moreover, hospital environment samples (such as basin, surface, and sewage) and patients’ screen samples were not included in our study, which would have provided more information on clonal dissemination. However, our findings provide valuable information for understanding the molecular epidemiology of CRKP in a non-tertiary hospital in China. Of note, most surveillance studies were performed in tertiary hospitals in mainland China. This study highlights the need for improvement of infection control practices in non-tertiary hospitals, as well as indicates the importance of performing regional resistance surveillance.
5.1. Conclusions
CRKP strains were mainly isolated from ICU and neurosurgery departments. Several chains of dissemination of CRKP were discovered in the hospital and region, as ST11 was the main epidemic clone. Multiple carbapenemase genes were detected, with blaKPC-2 being the most frequent. The resistance rate of CRKP strains to CAZ/AVI was higher in our study than in previous studies. Our findings highlight the need for more effective antimicrobial stewardship and infection control practices in non-tertiary hospitals in China.