1. Background
Klebsiella pneumoniae is one of the most common Gram-negative bacteria in the Enterobacteriaceae family, which usually causes the infection of the respiratory system and other parts of the body (1). Generally, K. pneumoniae widely exists on the surface of any object in nature and hospital environment and can be colonized on human skin, respiratory tract, and intestinal tract (2). When the immunity of the human body decreases, K. pneumoniae will cause several body infections, such as pneumonia, urinary tract infection, and meningitis (3, 4). When K. pneumoniae enters the blood, it will cause bacteremia and even sepsis, which will seriously threaten the lives of patients (5).
Antimicrobials are effective treatments for bacterial infections. However, the emergence of multidrug-resistant (MDR) bacteria has brought great challenges to clinical antimicrobial treatment, making bacteria resistant to most antimicrobials, such as β-lactams, aminoglycosides, fluoroquinolones, and even carbapenems (6). Klebsiella pneumoniae began to show resistance to aminoglycosides in the 1970s, followed by resistance to third-generation cephalosporins in the 1980s and 1990s and resistance to carbapenems in recent years (7). Carbapenems are the last resort for K. pneumoniae producing extended-spectrum β-lactamases (ESBLs) or MDR pathogens (8). However, when K. pneumoniae produces carbapenemase, carbapenems have no effect at all, and only other drugs, such as colistin and ceftazidime/avibactam, can be selected (9, 10).
Drug resistance caused by carbapenem-resistant Enterobacteriaceae (CRE) has become a serious public health problem worldwide (11). Different CRE can produce different carbapenemases. Carbapenemase is one of the β-lactamases, which has numerous types, and its main function is to hydrolyze carbapenems and other β-lactam drugs (12). According to the Ambler classification, carbapenemases can be divided into three categories (13). They are class A serine carbapenemases, class B metallo-β-lactamases (MBLs), and class D oxacillinase-type carbapenemases (OXA). Class A mainly includes types of KPC, IMI, GES, SME, and others. Class B includes types of NDM, Verona integron-encoded metallo-β-lactamase (VIM), IMP, GIM, SPM, and others. Class D mainly includes OXA-48, OXA-23, and others (14).
Klebsiella pneumoniae producing KPC carbapenemase was first detected in 1996 in the United States and then spread worldwide (15). Because blaKPC clonal spread includes horizontal transfer and plasmid, the transmission is fast and wide (16). At present, KPC carbapenemase is mostly produced by carbapenem-resistant K. pneumoniae (CRKP) isolated in China (17). Klebsiella pneumoniae producing VIM carbapenemase was first reported in 2007 in Spain (18). A previous study confirmed that the VIM gene was the most frequent gene of MBLs gene in the world (19).
There are numerous methods to detect the phenotype and genotype of carbapenemase, including the modified Hodge test, Carba NP test (20), modified carbapenem inactivation method (mCIM) combined with EDTA-CIM (21), colloidal gold (22), GeneXpert test (23), and polymerase chain reaction (PCR) (24). As a new identification method of carbapenemase phenotype, colloidal gold is simple and accurate. The PCR is a gold standard method; however, due to complicated operations, it is not available in laboratories to verify each isolate. Although there were numerous reports on carbapenemase produced by K. pneumoniae in China, and the present research team has relevant studies on carbapenemase, there was no report on the distribution of carbapenemase type of K. pneumoniae with blood infections in southern Anhui province, China.
2. Objectives
The main purpose of this study was to determine the prevalence of carbapenemase type of K. pneumoniae with blood infection isolated from blood culture samples.
3. Methods
3.1. Sample Origin
This retrospective study was carried out at Yijishan Hospital of Wannan Medical College, Wuhu, Anhui province, China, within September 2020 and August 2022. A total of 9255 blood culture samples from inpatients were processed in this study. All the samples were transported to the laboratory without leakage and pollution. All the patients met the requirements for clinical blood culture and signed informed consent. This study complied with the Declaration of Helsinki and was approved by the Institutional Review Board of Yijishan Hospital of Wannan Medical College.
3.2. Sample Culture and Strain Identification
All the blood culture samples were incubated in BACT/ALERT® 3D automatic instrument (bioMérieux, France) for at least 7 days. The positive samples were inoculated on blood agar, chocolate agar, and MacConkey agar. Then, the isolates were identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOFMS) (bioMérieux, France).
3.3. Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) method was used for the sensitivity and resistance of bacteria to antimicrobials. The MIC method was carried out by VITEK-2 automated instrument (bioMérieux, France). Carbapenems (i.e., ertapenem, imipenem, and meropenem) were determined to judge carbapenemase production. The interpretation of strain resistance to carbapenems was according to the Clinical and Laboratory Standards Institute (CLSI) M100-S30 guidelines.
3.4. Phenotype Detection of Carbapenemase
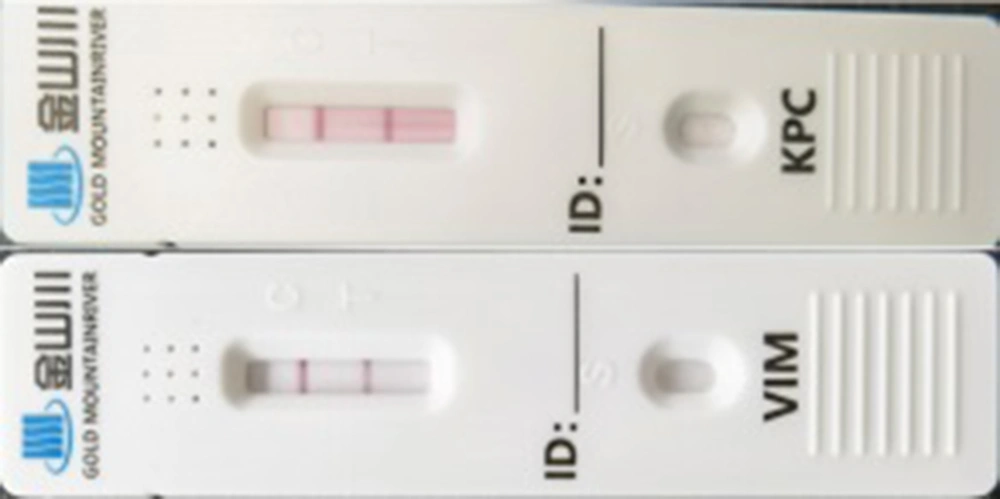
The colloidal gold (Era Biology, Tianjin) method was used to confirm the phenotype of carbapenemase produced in isolates. This technique was used to combine the sample with the gold-labeled antibody to form an antigen-antibody immune complex. The complex reacts with the coated and solidified carbapenemase monoclonal antibody through capillarity to form a double antibody sandwich detection strip on cellulose membrane, forming KPC, NDM, IMP, VIM, and OXA-48 corresponding detection lines (25).
3.5. Deoxyribonucleic Acid Extraction and Amplification of Carbapenemase Genes by Polymerase Chain Reaction
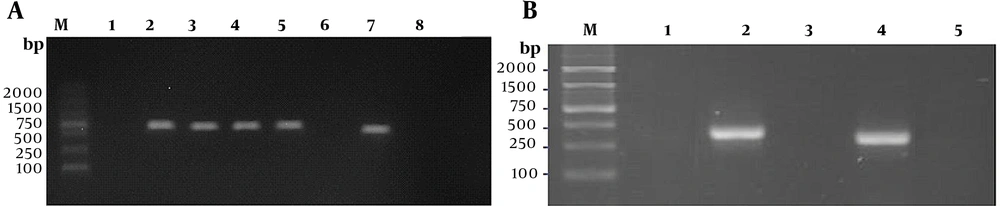
Crude plasmid deoxyribonucleic acid (DNA) was extracted using the alkaline denaturation method. The plasmid was suspended in TE buffer and stored at -20°C. The carbapenemase genes (blaKPC and blaVIM) were amplified by PCR. The primers were synthesized according to the sequence provided in the literature (Table 1) (26). A total of 15 μL reaction mixture includes 2.0 μL DNA template, 0.5 μL forward primer, 0.5 μL reverse primer, 7.5 μL 2 × PCR mixture, and 4.5 μL ddH2O. The PCR entailed the processing of the samples for initial denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, annealing at 56°C for 30 seconds, extension at 72°C for 60 seconds, and final extension at 72°C for 6 minutes.
| Gene | Primer Sequence | Product Size (bp) |
|---|---|---|
| KPC | 798 | |
| F | 5’-CGTCTAGTTCTGCTGTCTTG-3’ | |
| R | 5’-CTTGTCATCCTTGTTAGGCG-3’ | |
| VIM | 390 | |
| F | 5’-GATGGTGTTTGGTCGCATA-3’ | |
| R | 5’-CGAATGCGCAGCACCAG-3’ |
3.6. Validation of Polymerase Chain Reaction Products by Gel Electrophoresis
First, 2% agarose gel was prepared and poured into the mold. Then, the comb was inserted at an appropriate position and solidified at room temperature for 40 minutes. Then, 10 μL PCR product containing loading buffer was added to each hole, and electrophoresis was performed at 110 V for 30 minutes. Finally, gel images were collected under ultraviolet light.
3.7. Homologous Analysis of Strains
The isolates of CRKP were coated on the target plate, and 1 μL IVD HCCA matrix solution was added to the surface of the bacteria. After drying the sample, the target plate was sent into MALDI-TOFMS to collect the protein fingerprint peak map. The results were analyzed by MALDI-TOFMS SARAMIS software (version 4.1) and imported into the map database for cluster analysis. Those strains whose similarity was less than 70% were considered different types.
3.8. Quality Control
Strict operations were followed at each step. Bacterial identification and antimicrobial susceptibility testing (AST) were strictly in accordance with CLSI standards. During AST, K. pneumoniae ATCC BAA-1705 and BAA-1706 were used as control strains producing KPC carbapenemase and non-producing carbapenemase separately. In PCR reaction, K. pneumoniae carrying blaKPC or blaVIM gene was used as a positive control, and K. pneumoniae without any carbapenemase gene was used as a negative control.
4. Results
4.1. Source of Blood Samples
A total of 9255 samples were collected. The top five departments were the Department of Infectious Diseases, Department of Pediatrics, Department of Hematology, Intensive Care Unit, and Department of Nephrology of Yijishan Hospital, accounting for 20.96%, 16.74%, 16.69%, 14.33%, and 3.13% of the total blood culture, respectively.
4.2. Distribution of Isolated Strains
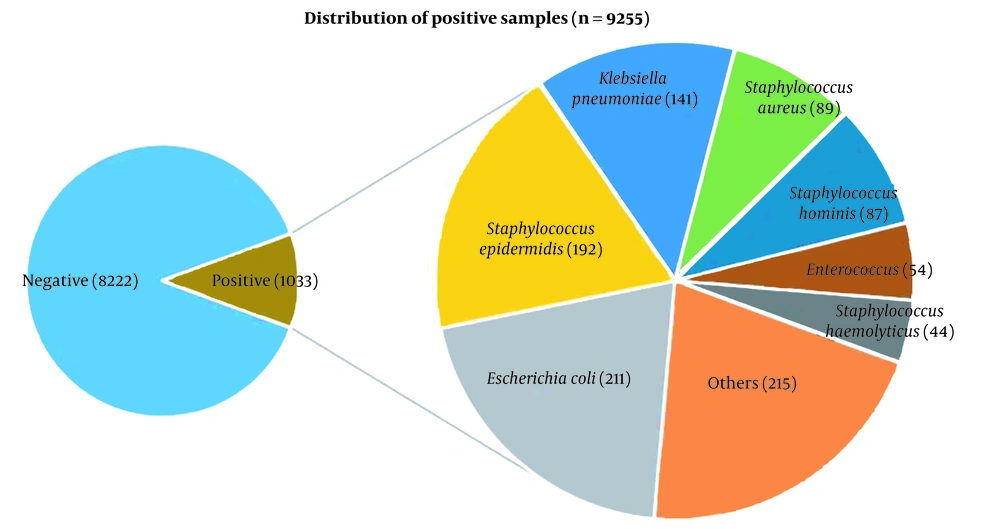
Among 1033 bacterial isolates, Escherichia coli was reported with the highest detection rate (20.43%, 211/1033), followed by Staphylococcus epidermidis (18.59%, 192/1033), K. pneumoniae (13.65%, 141/1033), and S. aureus (8.62%, 89/1033) (Figure 1).
4.3. Antimicrobial Susceptibility Testing Results of Klebsiella pneumoniae
Out of 141 strains of K. pneumoniae, only 11 isolates showed resistance to carbapenems. The AST results showed that 11 CRKP isolates were resistant to most antibiotics, including aztreonam, ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, ceftazidime, cefepime, cefoperazone/sulbactam, ciprofloxacin, levofloxacin, ertapenem, imipenem, and meropenem, but with high sensitivity to colistin (100%, 11/11), ceftazidime/avibactam (90.91%, 10/11), tigecycline (81.82%, 9/11), and amikacin (63.64%, 7/11) (Table 2).
| Antibiotics | Sensitive | Resistant |
|---|---|---|
| Aztreonam | 0 (0) | 11 (100) |
| Ampicillin | 0 (0) | 11 (100) |
| Ampicillin/sulbactam | 0 (0) | 11 (100) |
| Piperacillin/tazobactam | 0 (0) | 11 (100) |
| Ceftazidime | 0 (0) | 11 (100) |
| Cefepime | 0 (0) | 11 (100) |
| Cefoperazone/sulbactam | 0 (0) | 11 (100) |
| Ciprofloxacin | 0 (0) | 11 (100) |
| Levofloxacin | 2 (18.18) | 9 (81.82) |
| Ertapenem | 0 (0) | 11 (100) |
| Imipenem | 0 (0) | 11 (100) |
| Meropenem | 0 (0) | 11 (100) |
| Gentamicin | 1 (9.09) | 10 (90.91) |
| Amikacin | 7 (63.64) | 4 (36.36) |
| Tigecycline | 9 (81.82) | 2 (18.18) |
| Ceftazidime/avibactam | 10 (90.91) | 1 (9.09) |
| Colistin | 11 (100) | 0 (0) |
a Values are expressed as No. (%).
4.4. Phenotype Identification of Carbapenemase
Out of 11 CRKP isolates, 9 (81.82%) strains were observed to produce carbapenemase by the colloidal gold method. Among the aforementioned 9 isolates, 8 strains produced KPC carbapenemase, and 1 strain produced VIM carbapenemase (Figure 2).
4.5. Genotype Confirmation of Carbapenemase
All CRKP isolates underwent molecular screening for the detection of carbapenem resistance-related genes. The results showed that nine strains carried carbapenemase resistance-related genes, among which 88.89% (8/9) and 11.11% (1/9) of the isolates were found positive for blaKPC and blaVIM genes, respectively (Figure 3). The results were consistent with those of phenotype identification.
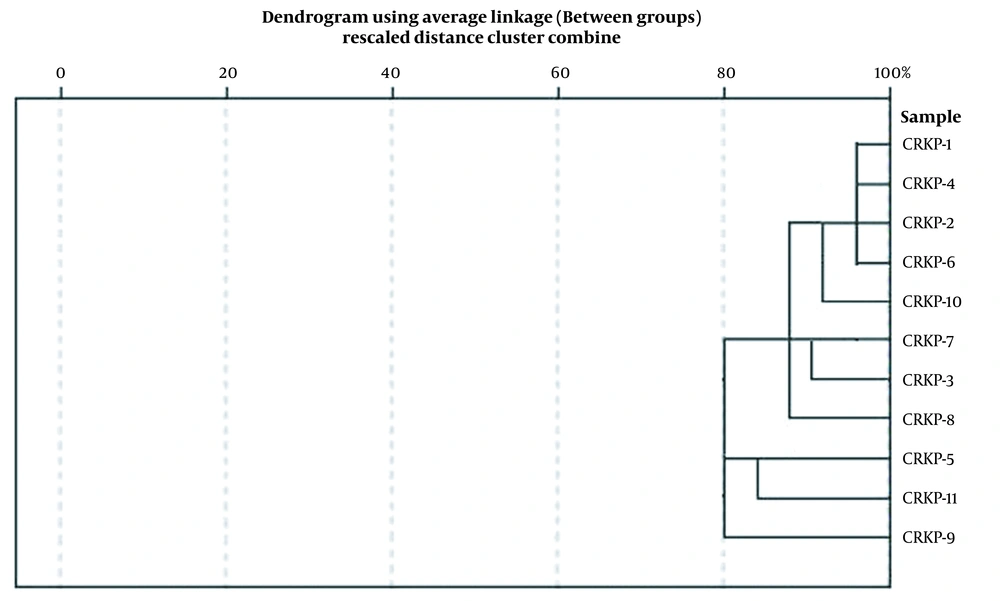
4.6. Homologous Analysis of Carbapenem-resistant Klebsiella pneumoniae
The cluster analysis and the dendrogram of CRKP by MALDI-TOFMS showed that the similarity of 11 CRKP isolates was more than 80%, indicating that they were closely related (Figure 4).
5. Discussion
Bloodstream infections (BSIs) caused by Enterobacteriaceae are serious threats to the lives of patients, resulting in a mortality rate as high as 48% (27). Carbapenems are one of the most effective treatments for BSIs. However, with the emergence and increase of bacteria producing carbapenemase, the resistance rate of carbapenems is increasing gradually (28). In hospitals, carbapenemase-encoding plasmids can be transferred among different Enterobacteriaceae through horizontal gene transfer and disseminated (16). As a result, CRE has spread worldwide, and few effective treatments are available (29). Klebsiella pneumoniae is one of the common pathogens in nosocomial infections, which can cause pneumonia, urinary tract infection, soft tissue infection, and even septicemia. In China, the infection caused by CRKP accounts for about 64% of that by CRE; nevertheless, the proportion varies among provinces or regions (30). The current study was designed to determine the proportion of K. pneumoniae in BSIs and carbapenemase type produced by CRKP in the south of Anhui province.
In this study, BSIs caused by K. pneumoniae accounted for 13.65% of total BSIs. Among all strains of K. pneumoniae, CRKP accounted for 7.80%. The AST of CRKP showed higher resistance to carbapenems which was comparable to the results of some previous studies from India (31), Turkey (32), and Malaysia (33). Generally, the effectiveness of one drug can be enhanced when used combined with another drug, even when the bacteria resist this kind of drug. Therefore, a multidrug combination can be regarded as an effective measure in the treatment of CRKP (34). Additionally, the present study showed that colistin, ceftazidime/avibactam, and tigecycline demonstrated high sensitivity to CRKP. Among them, all CRKP isolates were sensitive to colistin, 90.91% of CRKP isolates were sensitive to ceftazidime/avibactam, and 81.82% of CRKP isolates were sensitive to tigecycline. However, it has been reported that when tigecycline is used to treat CRKP, it will induce the strain to be resistant to tigecycline (35). The reason might be that the reduced sensitivity of CRKP is the role of RamA on the expression of the efflux pump AcrAB (36). Therefore, it is necessary to be cautious about tigecycline resistance when it is used clinically.
Recently, ceftazidime/avibactam has been a new β-lactamase inhibitor for the treatment of CRKP, especially for K. pneumoniae producing KPC carbapenemase (37). There have been numerous successful reports on the treatment of CRKP with ceftazidime/avibactam; however, there were also a few cases of resistance (38). In this study, one CRKP isolate was observed to be resistant to ceftazidime/avibactam; nonetheless, its relevant mechanism was not implemented. A previous study by the current research team demonstrated that the deletion of the outer membrane protein OmpK36 could lead to resistance to ceftazidime/avibactam in CRKP. Therefore, the cause of resistance can be further verified in this study.
The rapid identification of strains producing carbapenemase is important to ensure early specific treatment and the implementation of the most reasonable infection control measures. Recently, the application of some new diagnostic technologies has accelerated the identification of bacteria, such as the Carba NP test, rapid colloidal gold immunochromatography, MALDI-TOFMS (39), and molecular biology-based assays. Among the aforementioned tests, the colloidal gold method is simple and fast, and the results are highly consistent with the gold-standard method, which can be popularized in daily work. Although real-time PCR is a gold-standard method for the detection of carbapenemase encoding genes, it is inconvenient and expensive, and it is only used as a validation test. Carbapenemases are mainly divided into three categories; KPC is the representative of class A serine carbapenemase; NDM and VIM are common MBLs; OXA is mainly class D carbapenemase.
Numerous studies have shown that CRKP produces KPC at most, followed by NDM, IMP, VIM, and others. The current study proved that CRKP mainly produced KPC (88.89%), followed by VIM (11.11%); however, no other carbapenemase was detected, which was also verified by real-time PCR. Meanwhile, MALDI-TOFMS showed that these CRKP strains had high homology. This carbapenemase-type prevalence was different from previous reports in Greece (40), Iran (41), and India (42). It might be caused by the insufficient sample size of CRKP. This experiment can be repeated to further extend the research time and cooperation with other hospitals in Anhui province to better understand the current situation and drug resistance of CRKP in BSIs in Anhui province.
5.1. Conclusions
Carbapenemase produced by CRKP can lead to its resistance to carbapenems. The emergence and spread of genes, especially blaKPC and blaVIM, have threatened the treatment of CRKP. The colloidal gold method has the advantages of simplicity and rapidity, and PCR has the advantage of more specificity. A combination of the two methods for detection can aid in the accurate and early diagnosis and management of infectious diseases.