1. Background
Ulcerative colitis (UC) is known as a chronic immune-mediated disease, causing inflammation and ulcers in the colon (1). The first symptom of active disease is diarrhea mixed with blood. Many parts of the body outside the intestinal tract are affected by ulcerative colitis. In fact, the initiation of UC disease is in intestinal zones with high bacterial counts (2, 3). The pathogenesis of UC is complicated even though its cause is still unknown. Genetic background could be a risk factor for UC disease; however, microbiota and immune system may have roles in the occurrence of the disease (4). A wide range of bacterial species is involved in the inflammation of the colon, including Enterobacteriaceae, especially Escherichia coli (5). Escherichia coli strains living in the gut are not considered to be harmful. However, numerous studies have indicated that the number of adherent-invasive E. coli (AIEC) strains in inflammatory bowel disease (IBD) patients is significantly higher than previously estimated (6).
Adherent invasive E. coli can adhere to epithelial cells and invade cytoplasmic eukaryotic infectious cells owing to type 1 fimbriae (fimH), invasive plasmid antigen H (ipaH), and invasion-association locus (ial). Also, it can replicate into macrophages. Accordingly, it should be considered a separate pathogenic cohort triggering intestinal disease in human. Subsequently, studies proposed that AIEC can be associated with pathogenicity of IBD (7). In adherent-invasive E. coli, the fimH gene, in particular, helps in adhering and colonizing epithelial cells.
The ability to invade intestinal cells is acquired by ipaH and ial genes. The ipaH gene exists in multiple copies located on both chromosomes, and the plasmid is in charge of release in epithelial cells (8). Recently, there are no data about the prevalence of adherent-invasive E. coli in the large bowel of patients with inflammation (9). Adherent invasive E. coli pathotype has been gradually involved in the etiopathogenesis of ulcerative colitis (10). A number of culture-based and molecular-based studies support the theory that adherent-invasive E. coli especially those with fimH gene are a microbiological factor effective in IBD (11-13).
2. Objectives
The main aim of the present study was to determine the prevalence of adherent-invasive E. coli with fimH gene isolated from Iranian patients with ulcerative colitis.
3. Methods
3.1. Patients and Tissue Samples
In the present study, a group of biopsy samples of 60 subjects comprising 30 UC patients (12 males, 18 females, median age 36.9 years, age range 16 - 75 years) and 30 individuals without IBD (14 males, 16 females, median age 48.31 years, age range 19 - 76 years) were taken during colonoscopy. The study was approved by the ethics committee of Tehran University Medical Sciences.
3.2. Microbial Identification
Samples obtained during colonoscopy were transferred immediately into sterile vials containing either thioglycolate broth or saline (Sigma-Aldrich, Hi Media) and stored at -20°C. The biopsy specimens were homogenized and inoculated into Hi Chrome E. coli agar (Sigma-Aldrich, Hi Media) and incubated for 18 - 24 hours at 35 ± 2°C. The bacteria were stored in TSB broth containing 30% glycerol at -70°C until further analysis.
3.3. DNA Extraction from Biopsies for PCR
Biopsies were crushed and DNA was extracted by RTP® Mycobacteria kit (Berlin, Germany).
3.4. DNA Extraction from Culture for PCR
Isolated bacteria were prepared for PCR amplification. The bacterial colonies were harvested and centrifuged. The sediments were suspended in 500 µL of sterile deionized water, and boiled for 10 minutes. After centrifugation of the boiled samples at 19000 g for 5 minutes, the supernatant was used as DNA template in PCR assay (14).
3.5. PCR Assay
All isolates were tested for the occurrence of ipaH, ial, and fimH genes using PCR. The nucleotide sequence of primers (Macrogen, Pishgam) and size of product (base pairs) for amplification of fimH, ipaH, and ial genes are displayed in Table 1 (15, 16). PCR was performed in 25 μL solution comprising 5 μL of master mix (Amplicon, Pishgam), 1 μL of each primer (10 pm/μL), 2 μL of DNA template (50 ng), and 16 μL of ddH2O. Subsequently, the following thermal cycling conditions were used: 5 minutes at 94°C and 36 cycles of amplification consisting of 30 seconds at 95°C, 30 seconds at 56°C, and 1 minutes at 72°C, with 5 minutes at 72°C for final extension. PCR products were investigated by electrophoresis on a 1% agarose gel in 1X TBE buffer [10.8 g Tris and 5.5 g Boric acid, 0.5 M Na2EDTA (pH 8.0)] (17).
| Primers | Nucleotide Sequences (5’ - 3’) | Size of Product, bp |
|---|---|---|
| fimH | TATGGCGGCGTGTTATCTAG | 150 |
| CACAGGCGTCAAATAAAGCG | ||
| ipaH | GTTCCTTGACCGCCTTTCCGATACCGTC | 619 |
| GCCGGTCAGCCACCCTCTGAGAGTAC | ||
| ial | CTGGATGGTATGGTGAGG | 320 |
| GGAGGCCAACAATTATTTCC |
Primers and Annealing Temperature for Amplification of the AIEC Genes
3.6. Statistical Analysis
The data were evaluated by Pearson Chi-Square test. A P value < 0.05 was considered statistically significant.
4. Results
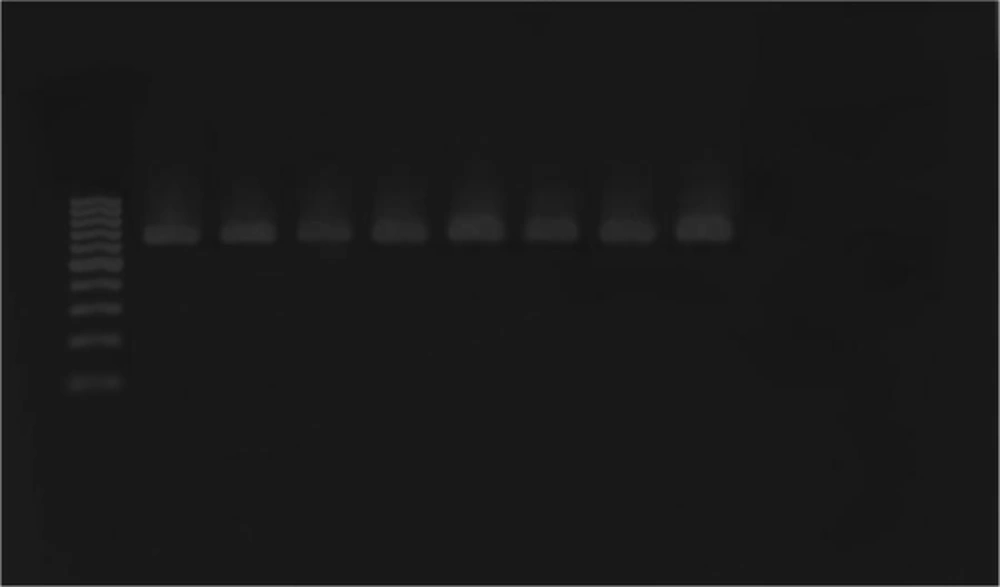
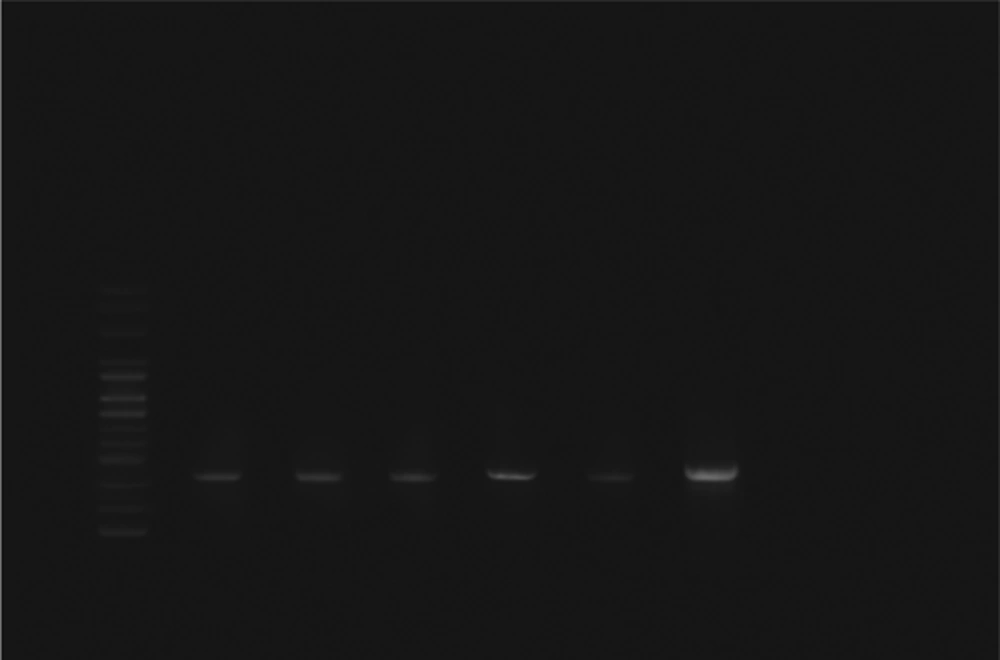
Escherichia coli strains were detected from biopsy samples of 24 patients with UC (80%) and 26 healthy controls (86.7%). All isolated bacteria were confirmed as AIEC by biochemical tests and PCR assay. The ipaH, ial, and fimH genes were amplified by utilizing particular primers and became visible at approximate bands of 619, 320, and 150 bp on polyacrylamide gel, respectively (Figures 1, 2 and 3). Among isolated bacteria, the presence of fimH was confirmed in 53.3% (n = 16) of specimens from UC patients, while 46.7% (n = 14) were negative. Also, this gene was detected in 4 control subjects (13.3%) while the remaining 86.7% (n = 26) lacked this gene (Table 2). In addition, amongst 60 biopsy samples taken during colonoscopy, the fimH gene was detected in 17 (56.7%) patients with UC and 3 (10%) control subjects (Figure 3). Accordingly, 43.3% (n = 13) of UC patients and 90% (n = 27) of control subjects did not yield any amplicon in PCR assay (Table 3). Therefore, fimH gene in E. coli strains isolated from UC patients was more frequent than that of control population in PCR assay. Moreover, PCR assay was more reliable than cultivation. Based on the results, the association of fimH gene presence in adherent-invasive E. coli with UC patients was statistically significant (P < 0.05). Also, all positive amplified fragments were sequenced.
| Population | fimH | ||
|---|---|---|---|
| Positive, No.% | Negative, No.% | P Value | |
| UC, No. = 30 | 16 (53.3%) | 14 (46.7%) | 0.001 |
| Control, No. = 30 | 4 (13.3%) | 26 (86.7%) | 0.001 |
The Presence of the fimH Gene of AIEC in Isolated from Two Groups (UC Patients and Control Subjects) by PCR Assay
| Population | fimH | ||
|---|---|---|---|
| Positive, N.% | Negative, N.% | P Value | |
| UC, No. = 30 | 17 (56.7%) | 13 (43.3%) | 0.000 |
| Control, No. = 30 | 3 (10.0%) | 27 (90.0%) | 0.000 |
The Presence of the fimH Gene of AIEC in Biopsy Samples from Two Groups (UC Patients and Control Populations) by PCR Assay
5. Discussion
The link between ulcerative colitis disease and the presence of adherent-invasive E. coli carrying fimH gene has been studied in recent years. The pattern of epidemiological characteristics and risk factors related to geographical and regional variations are still unknown (18-20). This study was carried out to determine the prevalence and possible role of AIEC with fimH gene in Iranian patients with UC by PCR assay since limited data are currently available on this subject. To clarify the possible colonization of AIEC in intestinal tract, we screened tissue samples for ipaH and ial genes by PCR assay as these genes have been reported in many cases to be involve in AIEC invasiveness (21). However, there are other studies proposing that these genes are significant virulence determinants in many extra intestinal infections in human, especially infections of urinary tract caused by E. coli (22).
Most studies indicate that modified microbial components and function in IBD can lead to the increased immune response. However, genetic, phenotyping, and microbial diversity within UC patients indicate that this disease is the heterogeneous cohort of different disorders, which probably has predictable natural histories (23). However, recent studies demonstrated that Entrobacteriaceae, E. coli, and Mycobacterium paratuberculosis strains are associated with IBD (24). The current study showed that there was a higher rate of fimH gene in UC patients than control subjects; a difference that was statistically significant. Despite the results of this study, some investigators did not observe any increase in AIEC among UC patients (25, 26).
In this regard, Raso et al. reported that AIEC was not detected in patients with UC and control subjects (27) and likewise, other studies found a higher prevalence for fimbriae I, encoded by the gene fimA, in UC patients compared to healthy individuals (28, 29). However, in our study, 56.7% of the patients with UC were positive for AIEC, in particular for those with fimHgen; a rate that was significantly higher than the rates reported by previous studies. Martin et al. indicated that E. coli is more common in patients with IBD in comparison with control people (30). It can be inferred that fimH gene of AIEC may trigger UC disease. But the absence of fimH gene in AIEC isolated from UC patients may indicate the point that other bacterial genera may also involve in this disease.
In many cases, several studies indicated that invasive E. coli strains are associated with Crohn’s disease (31). Based on previous studies, AIEC strains were identified as true invasive strains involved in the pathogenesis of IBD. The invasive ability of AIEC, acquired by fimH gene, helps its translocation to pass human intestinal barriers and move inside deep tissues (32, 33). In spite of the interesting debate, with the exception of assessing AIEC strains with fimH gene, this gene has been obtained in other strains of E. coli. It can be present in 68% of uropathogenic E. coli (UPEC) strains which are pathogenic in urinary tract (34). The evidence gathered proposes that adhesive and invasive E. coli strains could be involved in ulcerative colitis pathogenesis.
Also, limited data concerning the relation of adherent-invasive E. coli and fimHgen with UC disease in Iran are available. Therefore, we evaluated the presence of fimH gene of AIEC and its relation with UC disease. This study suggests that there is a possible role for this bacterium in pathogenesis of UC disease.
5.1. Conclusion
Several factors have been implicated in the progression of inflammatory bowel disease. In the present study, detection of AIEC with fimH gene from UC patients demonstrated that AIEC with fimH may be a predisposing factor in UC patients. This study provides the first clinical investigation about the relationship between AEIC, especially those with fimH gene, with ulcerative colitis disease in Tehran, Iran.