1. Background
Listeria monocytogenes (LM) is an intracellular Gram-positive pathogen causing listeriosis in both humans and various animal species (1). As a ubiquitous food-borne bacterium, LM can infect the host through contaminated food or drinking water. During the process of invasion, LM can express and deploy a variety of virulence factors, thereby breaking through the blood-intestinal, blood-brain, or placental barriers to cause meningitis, miscarriage, and sepsis in humans (2, 3). Since LM poses a great threat to food safety, it has been classified as one of the most important food-borne bacteria by the World Health Organization (WHO) (4). Bacterial small RNA (sRNA) is a class of non-coding RNAs that are usually transcribed within the intergenic region of the bacterial chromosome but do not encode proteins (5). In contrast to regulatory proteins, sRNA allows bacteria to respond rapidly to various environmental conditions during infection (6, 7).
In order to survive and proliferate in hosts, the pathogen can perceive changes in the host’s internal environment and regulate the expression of its virulence genes accordingly through various modulators and signaling cascades (8). Among regulatory repertoires, sRNA is now considered an important gene expression regulator at the post-transcriptional level. In recent years, many studies have reported that sRNA can be involved in cellular metabolism (9), physiological growth, and pathogenicity of bacteria through interacting with target mRNAs, thereby modulating responses to a variety of environmental stresses and facilitating survival in hosts (5). In 2011, Mraheil et al. identified about 150 sRNAs in LM (10) through sRNA transcriptomic analysis during the growth of the bacterium in macrophages. Among them, Rli82 was one of the identified sRNAs showing significant differential expression. Meanwhile, Rli82 deletion alters the growth pattern of LM; however, the regulatory function of Rli82 sRNA remains unknown so far.
2. Objectives
The main purpose of this study was to unveil the role of Rli82 sRNA on the motility and pathogenicity of LM and to further ascertain the potential regulatory mechanism of this sRNA. To achieve this goal, overlap extension PCR (SOE-PCR) and homologous recombination techniques were employed to construct Rli82-deleted and complementation LM strains. Then, the potential target gene regulated by Rli82 was predicted and verified to provide new insights into the mechanism of sRNA-mediated control of flagellum-related genes in LM.
3. Methods
3.1. Plasmids, Strains, and Culture Condition
The shuttle vectors of pKSV7 and pHT304 were used to generate Rli82-deleted and complementation LM strains. EGD-e strain was cultured in Brain Heart Infusion (BHI) Broth (Difco, USA) at 37°C, whereas Escherichia coli DH5α and BTH101 strains were cultured in LB (Difco, USA) medium at 37°C.
3.2. Primer Design
The specific primers used in this study were designed based on the LM EGD-e genome sequence deposited in GenBank (accession number: AL591824) using Primer Premier 5.0 software. Table 1 shows the detailed information of the designed primers.
| Primer Name | Primer Sequence (5’→3’) | Product Size (Bp) |
|---|---|---|
| F1 | GGTACCAAGACACCAGTTCCGTTTA | 495 |
| F2 | GTTCTGTTATACAGTATCTTTTTGATGACTAAAGTATATA | |
| F3 | TATATACTTTAGTCATCAAAAAGATACTGTATAACAGAAC | 316 |
| F4 | AAGCTTCCTATTAGAAACACGAGCATTA | |
| F5 | TGCTGTCTTACCAGTAGGCTCA | 1625 |
| F6 | AAGAAATCAGTGGAAGTAGCCC | |
| 82 F | GGATCCATCCTCCTATAGGCACTTTTTAGTATCTA | 70 |
| 82 R | AAGCTTATATACCGTACAGAATAACAAGAAGGTAC | |
| flaA-laczF | AAGCTTTTTGGACAACTTTTCTGTTCA | 251 |
| flaA-lacz R | GGTACCGTATTTACTTTCATTTGTGTTTCC | |
| flaA F | AACAAGCAACTGAAGCTATTGATGAATT | 247 |
| FlaA R | TGCGGTGTTTGGTTTGCTTGA | |
| 16sRrna F | CACTGGGACTGAGACACGG | 243 |
| 16sRrna R | GGACAACGCTTGCCACCTA | |
| FlaA F | GTCGGATCCATGAAAGTAAATACTAATAT | 864 |
| FlaA R | CATCTCGAGTTAGCTGTTAATTAATTGAGT | |
| GAPDH F | CGGGATCCATGACAGTTAAAGTTGGTATTAA | 1011 |
| GAPDH R | CCTCGAGTTATTTAGCGATTTTTGC |
3.3. Generation of Rli82 Gene-Deleted and Complementation Strains
Briefly, the LM EGD-e strain was cultured in BHI at 37°C for 12 h. The genomic DNA of LM was extracted according to the protocol of a bacterial genomic DNA extraction kit (Omega, USA). The upstream and downstream homology arms of the Rli82 gene were amplified using two pairs of primers (F1 - F2 and F3 - F4). These fragments were then used to generate Rli82-deleted mutant strain (ΔRli82) by SOE-PCR. Then, the ΔRli82 fragment was cloned into a pMD19-T simple vector (TaKaRa, Japan) to generate pMD19-T-ΔRli82. The pMD19-T-ΔRli82 and pKSV7 plasmids were double digested with Kpn I and Hind III (TaKaRa, Japan), and the target fragments were recovered and ligated with T4 DNA ligase (TaKaRa, Japan) at 16°C to produce the recombinant shuttle plasmid (pKSV7-ΔRli82). After that, pKSV7-ΔRli82 was transformed into LM EGD-e competent cells by electroporation (2500 V, 5.0 ms), and positive clones were screened by PCR using F5 - F6 primers.
The positive clones were passaged in BHI medium at a concentration of 10 μg/mL chloramphenicol for 15 generations at 42°C and in chloramphenicol-free BHI liquid medium for 15 generations at 30°C. The obtained recombinant LM-ΔRli82 was verified by PCR and sequencing. For the generation of the complementation strain, the Rli82 gene was amplified in LM EGD-e using 82F-82R primers and cloned into pHT304 plasmid to generate pHT304-Rli82. Then, pHT304-Rli82 was transformed into LM-ΔRli82 competent cells by electroporation, and the positive clones were screened on plates containing solid BHI at a concentration of 5 ug/mL of erythromycin. Positive clones were further verified by sequencing to obtain the complementation strain (LM-ΔRli82/Rli82) (11).
3.4. Determination of Motility
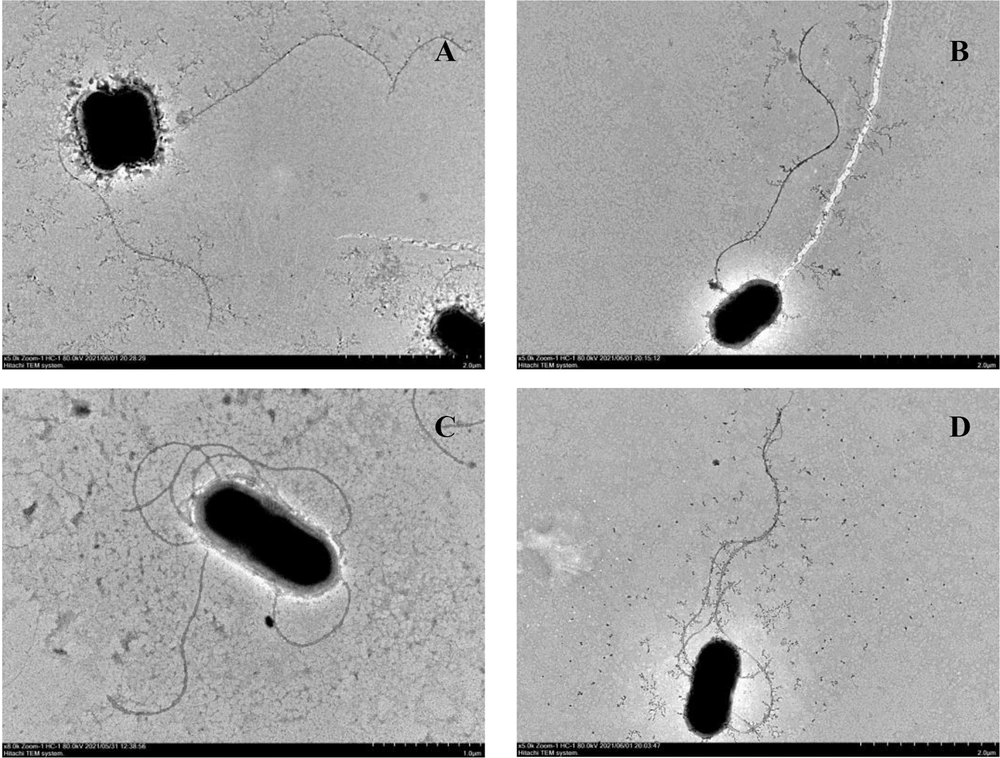
The motility of LM EGD-e, LM-ΔRli82, and LM-ΔRli82/Rli82 strains was assayed in the BHI semi-solid medium at 25°C. In brief, individual clones from these strains were harvested and washed three times with 0.01 M PBS buffer (pH = 7.2), centrifuged, and subjected to negative staining with 2% phosphotungstic acid solution (Sigma, USA). The morphological characteristics of these bacteria were observed using transmission electron microscopy (TEM) (HT7700, HITACHI, Japan). The flagella of 50 bacteria per strain were counted.
3.5. Determination of Pathogenicity
Mice were infected with LM EGD-e, LM-ΔRli82, and LM-ΔRli82/Rli82 by intraperitoneal injections. To determine LD50, bacterial concentration was adjusted to the same level (approximately 109 cfu/mL) for all three strains, from which a series of 10-fold dilutions to 105 CFU/mL were prepared. Then 6-8-week-old BALB/c mice were divided into 5 groups, and each mouse was injected intraperitoneally with 0.5 mL of the prepared bacterial solution, and 0.01 M PBS (pH = 7.2) was used as the control. After animal infection, mortality was monitored for 10 consecutive days in the study groups, and LD50 was calculated by the Spearman-Kärber method. In parallel, bacterial loads in the liver and spleen were determined in infected mice, and histopathological changes in these organs were observed after HE staining (11).
3.6. Target mRNA Prediction and Verification
The potential genes targeted by Rli82 were analyzed using TargetRNA2 bioinformatics online software (http://cs.wellesley.edu/~btjaden/TargetRNA2/). To verify the interaction between Rli82 and target genes, the two-plasmid reporter system based on E.coli (BTH101 strain) was employed. Briefly, the recombinant plasmids of pUT18C-Rli82 and pUT18C-ΔRli82 (Rli82 without the base-pairing region) and pMR-LacZ-target (the 5’-UTR region of flaA mRNA) vectors were constructed and co-transformed into E. coli BTH101 competent cells. Then, positive clones were cultured on LB agar containing X-gal and IPTG (TaKaRa, Japan) at 37°C for 12 h. Color change in the lawn solution was monitored, and the optical density (OD450 nm) of the lawn solution rinsed from the plates was determined.
3.7. Quantitative Real-time RT-PCR
Briefly, LM EGD-e, LM-ΔRli82, and LM-ΔRli82/Rli82 strains were incubated in the BHI medium at 25°C for 16 h, and total RNA was extracted using Trizol reagent in compliance with the instructions of the provider (Invitrogen, USA). Then, cDNA was synthesized using the AMV Reverse Transcription Kit (TaKaRa, Japan) following its instruction manual. Quantitative real-time RT-PCR (qRT-PCR) was performed on Light Cycler 480 (Roche, Switzerland) using a SYBR Premix Ex TaqTM kit (TaKaRa, Japan). The relative transcription levels of the target genes and Rli82 were calculated by the 2-ΔΔCT method (12). The 16s rRNA gene was employed as an internal reference control.
3.8. Western Blot Analysis
Western blot was performed as previously described (13). Briefly, bacterial protein was extracted using the Bacterial Protein Extraction Kit (Sangon Biotech, China) and analyzed by SDS-PAGE, followed by Western blot analysis using mouse-specific primary antibodies (1: 2000) and HRP-labeled rabbit anti-mouse IgG (Sigma, USA) secondary antibodies (1: 5000). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference protein. Image J software was applied to quantify the protein bands.
3.9. Statistical Analysis
SPSS software (SPSS Inc., Chicago, IL, USA) was used for the statistical analysis of the data. The data were presented in figures as mean values ± standard deviation (SD) from three independent experiments. The analysis of variance (ANOVA) was used to compare continuous variables, while the chi-square test was employed to analyze categorical variables. A P value < 0.05 was considered to be statistically significant, while P values < 0.01 were considered extremely significant.
4. Results
4.1. Generation of Rli82 Gene-Deleted and Complementation Strains
The deletion mutant (LM-ΔRli82) and complementation (LM-ΔRli82/Rli82) strains were successfully constructed and verified by PCR, restriction enzyme digestion, and sequencing (Appendix 1, Appendix 2, and Appendix 3, respectively).
4.2. Effects of Rli82 Gene Deletion on LM Motility
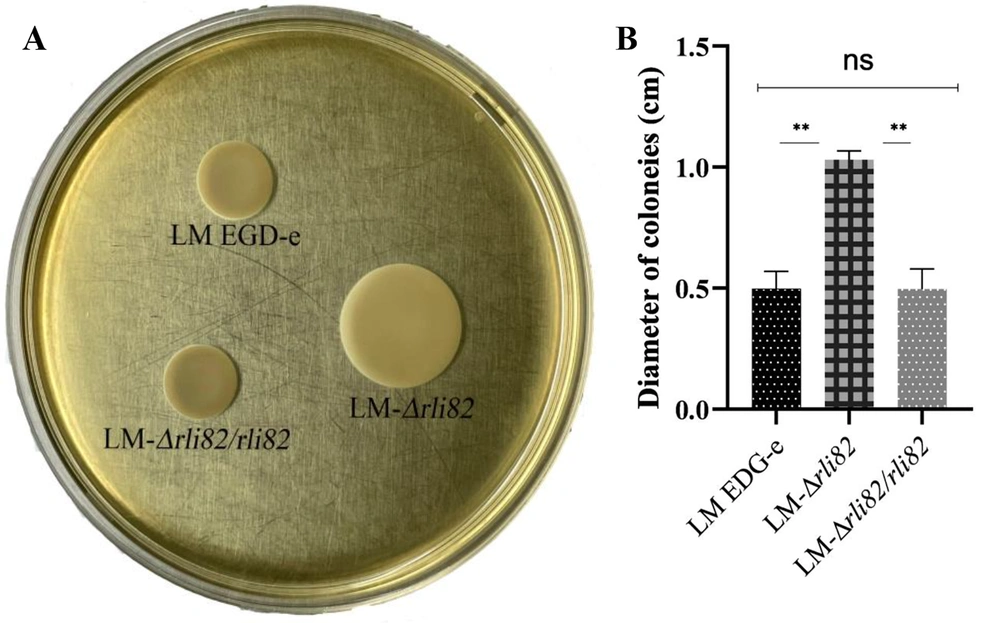
The mean diameter of the colonies formed by LM-ΔRli82 was significantly larger than that of LM EGD-e in semi-solid plates (Figure 1A and B), indicating that the motility of LM-ΔRli82 was significantly increased. Moreover, LM EGD-e bacteria carried 1.2 flagella per cell on average, whereas 4.3 flagella were counted on average for LM-ΔRli82, showing a significantly higher value in the mutant than in the parental strain (Figure 2).
4.3. Effects of Rli82 Gene Deletion on LM Pathogenicity
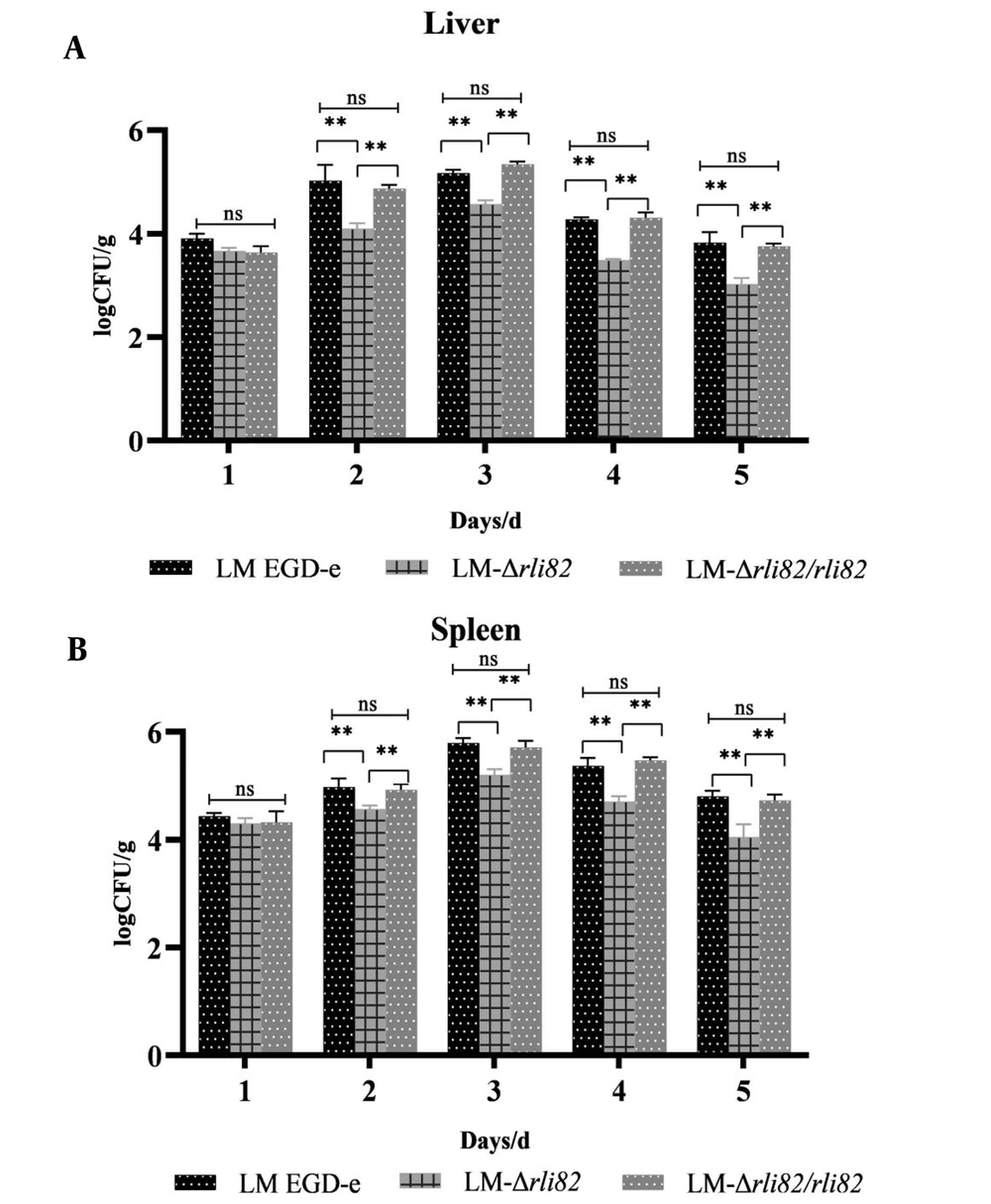
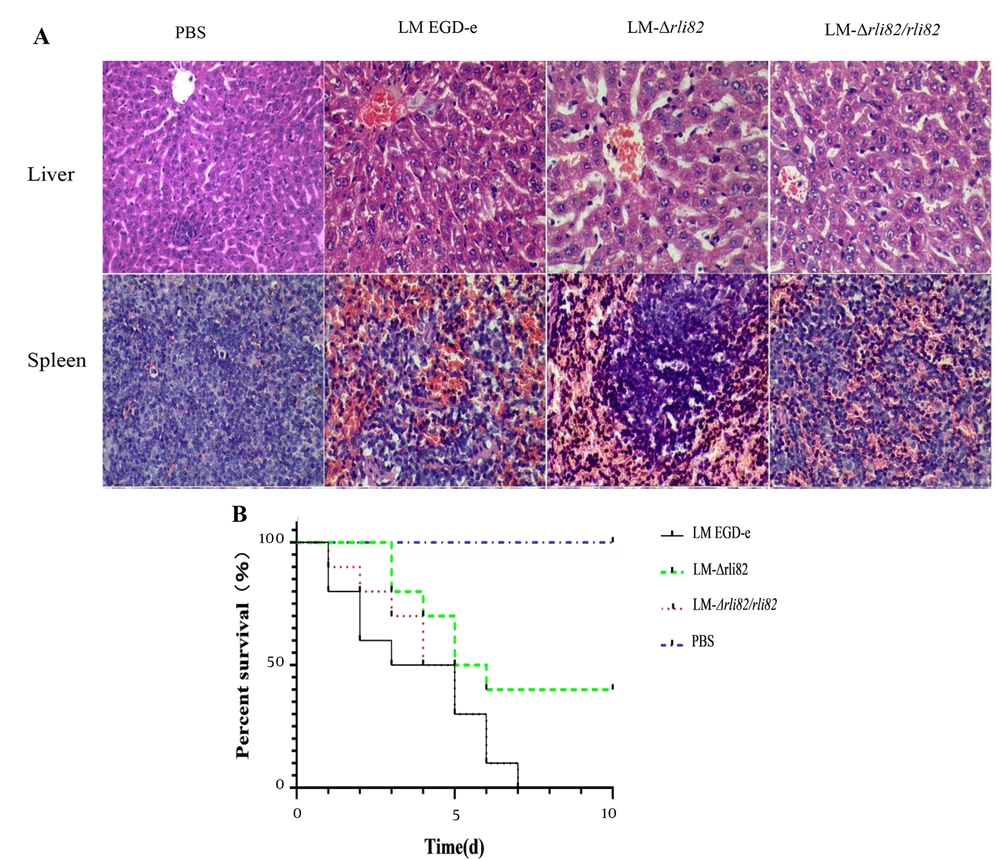
The LD50 of LM-ΔRli82 was 107.00 CFU/mL, which was significantly higher than that of LM EGD-e (105.56 CFU/mL) and LM-ΔRli82/Rli82 (105.95 CFU/mL) (Appendix 4), suggesting that the virulence of LM EGD-e was significantly decreased when the Rli82 gene was deleted. Moreover, bacterial loads in the liver and spleen of mice infected with LM-ΔRli82 were significantly lower than those of animals infected with LM EGD-e and LM-ΔRli82/Rli82 (Figures 3A and 4B). Histopathological changes in LM-infected mice revealed distinct necrotic foci, central venous congestion, and infiltration of inflammatory cells in the surrounding obstructive tissue in the liver. Moreover, spleeny nodules in the spleen were enlarged, and tissue congestion was evident. However, these pathological changes were significantly ameliorated in LM-ΔRli82 infected mice as compared to LM EGD-e and LM-ΔRli82/Rli82 infected animals (Figure 4A). LM-ΔRli82-infected mice survived significantly longer and had a significantly higher survival rate compared to those infected with LM EGD-e and LM-ΔRli82/Rli82 (Figure 4B), suggesting that sRNA Rli82 deficiency hampered the pathogenicity of LM.
4.4. Potential Target mRNAs Modulated by Rli82
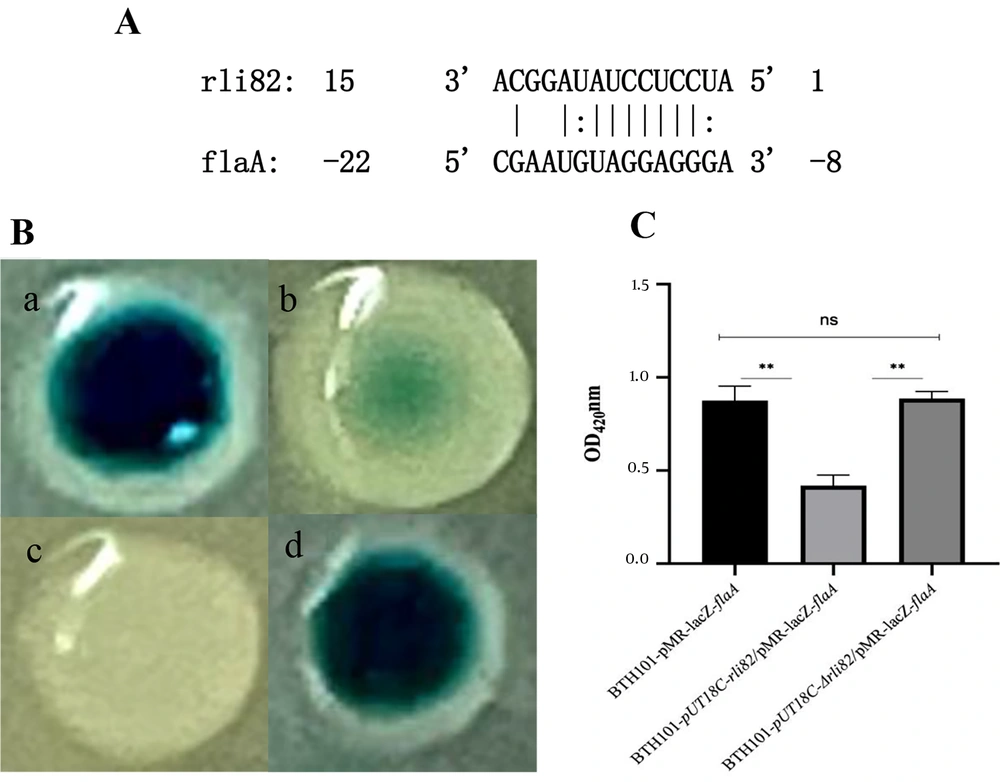
Bioinformatic analyses revealed that Rli82 was located at positions 910875 - 910944 on the genome of LM EGD-e (accession number: AL591824) (Appendix 5A). Regarding its secondary structure, Rli82 depicted a linear-shaped neck-loop structure with five loops and four complementary double strands (Appendix 5B). Online software, TargetRNA2, suggested flaA mRNA as a potential target for Rli82 based on a base-complementary segment (-22 ~ - 8 bases) in the 5’-UTR of the mRNA that paired with Rli82 (15~1 bases), implying that flaA mRNA could be potentially modulated by Rli82 at the post-transcriptional level (Figure 5A).
Prediction and verification of the target mRNA of Rli82. (A) The predicted target gene of Rli82; (B) Colonies of BTH101 (a): BTH101-pUT18C-Rli82/pMR-lacZ-flaA; (b): BTH101-pUT18C/pMR-lacZ-flaA; (c): BTH101-pUT18C/pMR-lacZ; (d): BTH101-pUT18C-ΔRli82/pMR-lacZ-flaA); (C): Comparison of OD at 450 nm of the flushing fluid between BTH101-pUT18C-Rli82/pMR-lacZ-flaA, BTH101-pUT18C/pMR-lacZ-flaA, and BTH101-pUT18C-ΔRli82/pMR-lacZ-flaA colonies
4.5. Verification of Interaction Between Rli82 and the Target mRNA
The two-plasmid reporter system based on E. coli showed that the bacterial strain co-transformed by pUT18C-Rli82 and pMR-LacZ-flaA formed deeper dark green colonies compared to the strain co-transformed by pUT18C and pMR-LacZ-flaA, accompanied by a significant 2-fold increase in the OD450nm of lawn’s flushing fluid. Meanwhile, there was a significant difference between the strain co-transformed by pUT18C-Rli82 and pMR-LacZ-flaA and the strain co-transformed by pUT18C-ΔRli82 and pMR-LacZ-flaA (Figure 5B and C). The results suggested that there was a substantial interaction between Rli82 and flaA mRNA.
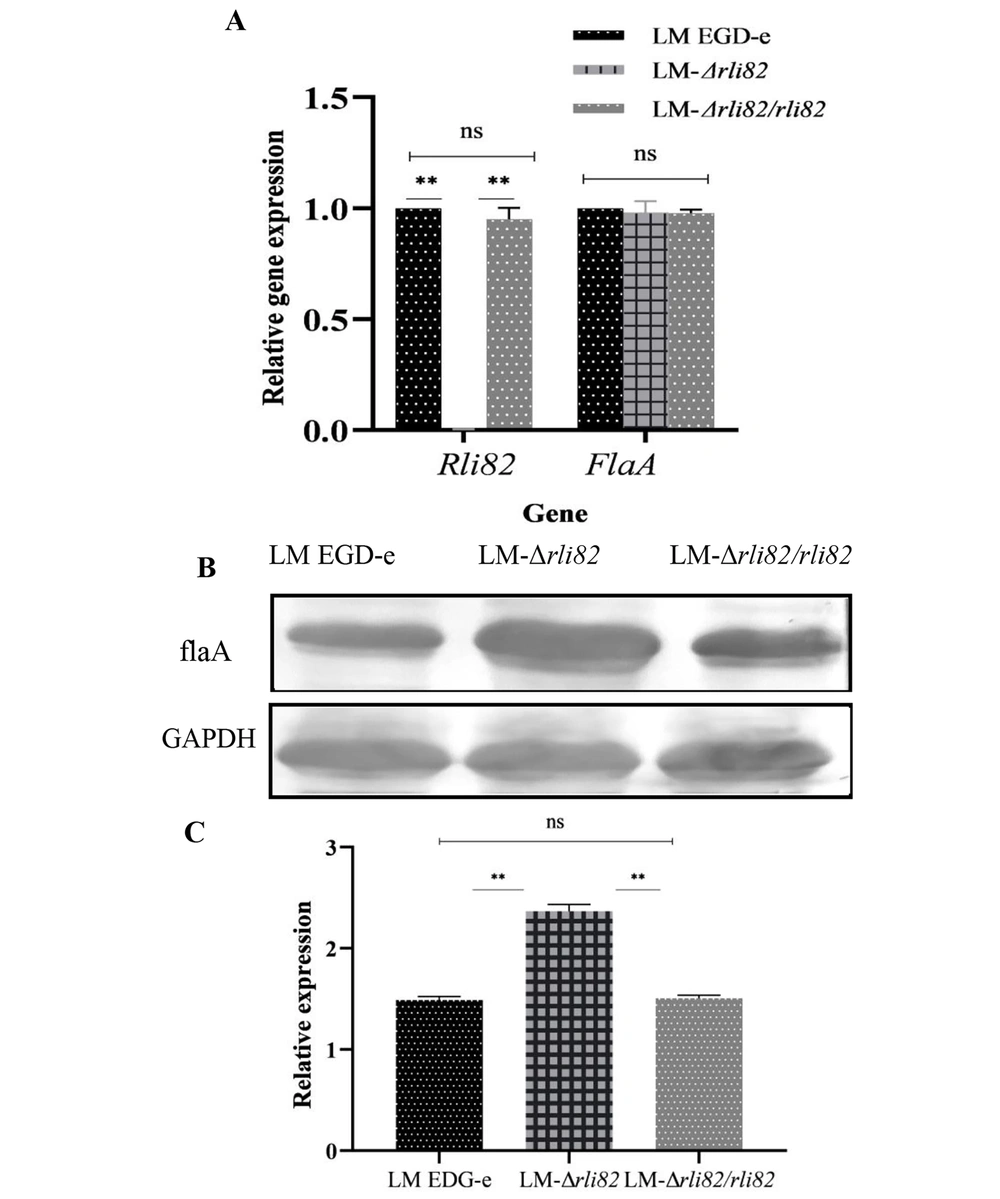
4.6. FlaA Gene Expression Analysis
Compared with LM EGD-e and LM-ΔRli82/Rli82 strains, the mRNA level of the flaA gene was not significantly elevated in the LM-ΔRli82 strain (P > 0.05). Meanwhile, there was no significant difference in the mRNA level of Rli82 between LM EGD-e and LM-ΔRli82/Rli82 (Figure 6A). However, it was revealed that the expression level of flaA protein was significantly higher in LM-ΔRli82 strain compared to LM EGD-e and LM-ΔRli82/Rli82 strains (Figure 6B and C), implying that Rli82 could negatively regulate the gene expression of flaA.
5. Discussion
So far, many studies have shown that sRNAs can act on target mRNAs to affect their transcriptional and translational levels (14, 15), whereby sRNAs can be involved in the regulation of the metabolism and virulence of bacteria (7, 9, 16). It is generally accepted that sRNAs regulate target genes’ mRNAs in a variety of ways. First, sRNA pairing at the Shine-Dalgarno (SD) region will suppress the binding of ribosomes with the mRNA, inhibiting the initiation of translation (17). In LM, sRNA LhrA inhibits the translation of lmo0850 by binding to its SD region (18). Second, sRNA can release the SD region, which, under normal conditions, is sequestered in a secondary structure, activating the translation of the target mRNA by ribosomes. In E. coli, sRNA Mcas can unlock the secondary structure of flhDC mRNA, thereby releasing the SD sequence and facilitating the translation process (19).
Alternatively, sRNAs may act on the far upstream of the ribosome binding site (RBS) of the target mRNA at its 5’-UTR, which protects the target mRNA from degradation by concealing its RNase E cleavage site. In this case, sRNA may promote the stability of the target mRNA and thus facilitate its translation (20). In Streptococcus, sRNA FasX in streptococci can bind to ska mRNA and prevent its degradation by RNase E, thereby stabilizing this mRNA and maintaining the translation of ska protein (21). Moreover, sRNA can also bind to a sequence near the ribosome binding site of the target mRNA, competing with 30S ribosomes for this binding site, resulting in the suppression of translation (22, 23). In Salmonella typhimurium, sRNA RyhB binds to fhlA mRNA and interferes with translation initiation (24). Herein, our experiments, combined with bioinformatics analyses, demonstrated that sRNA Rli82 might bind to the SD region of flaA mRNA, inhibiting the translation of FlaA protein, thus playing a vital role in the control of motility and pathogenicity of LM.
Previous studies have proven that the flagellum is closely related to physiological processes such as environmental stress tolerance, motility, and pathogenicity in bacteria (13, 25, 26). Existing studies have shown that the flagellum is composed of three parts, namely the flagellar filament, consisting of the flagellar subunit, hook, and basal body (27). Among these, the flagellar subunit is composed of flaA and other proteins (28). It has been shown that the deletion of the flaA gene can impair flagellar formation and interfere with the motility of LM (29). Here, target prediction analyses revealed that Rli82 was capable of complementary pairing with bases at the positions -22 to - 8 in the 5’-UTR of flaA mRNA, a site possibly representing the ribosomal binding site (RBS). Furthermore, the motility of the LM-ΔRli82 strain was significantly enhanced, which was in agreement with the observation of more flagella in LM-ΔRli82 than in LM EGD-e. Collectively, combined with the results of bioinformatic analyses, it can be noted that Rli82 may negatively modulate the expression of flaA mRNA by occupying its ribosomal binding site.
It has been proven that the functioning of LM flagella is restricted to temperatures below 37°C due to the opposing activities of the MogR transcriptional repressor and the GmaR anti-repressor (13, 30-32). Once LM enters the host, however, the biosynthesis of flagella is suppressed to help the bacterium evade the host’s immune system, thereby facilitating its survival and proliferation in vivo (31). Here, our results revealed that LM-ΔRli82 could produce more flagella than LM EGD-e and LM-ΔRli82/Rli82 at 25°C, suggesting a role for sRNA Rli82 in flagellar formation. Generally, LM maintains strong motility in the extracellular environment at temperatures below 37°C by enhancing the production of flagella, thereby expediting its chemotaxis, biofilm formation ability, and infectivity. However, the underlying mechanisms through which sRNA Rli82 can modulate flagella formation in LM need to be further elucidated by transcriptomic analyses.
5.1. Conclusions
Taken together, this study demonstrated that sRNA Rli82 was involved in the motility and pathogenicity of LM via modulating flaA mRNA at the post-transcriptional level. This observation provides new insights into sRNA-based modulation of the expression of flagella-related genes in LM.