1. Background
Klebsiella pneumoniae is a Gram-negative, oxidase-negative, non-spore-forming, facultatively anaerobic, and non-motile bacillus. It is enveloped by a characteristically thick polysaccharide capsule, which increases its resistance against many host defense mechanisms (1). Klebsiella pneumoniae isolates are abundantly found in nature and colonize the soil, surface waters, sewage, plant surfaces, and mucous surfaces of mammals. About one-third of the healthy population are intestinal carriers of this bacterium, making it the most common pathogenic species of Klebsiella (2). Klebsiella species are the third most common cause of hospital-acquired infections (HAIs) after Staphylococcus aureus and Clostridium difficile. Klebsiella pneumoniae is an opportunistic pathogen widely found in hospital environments and on medical devices. This bacterium mainly affects hospitalized patients with compromised immune systems. It has been isolated from various infections, such as pneumonia, sepsis, bacteremia, meningitis, urinary tract infections, and purulent abscesses in various organs, especially the liver (2).
Hospital-acquired pneumonia caused by K. pneumoniae may occur within 48 hours after hospital admission (3). The mortality rates of K. pneumoniae infections have been reported at 15% - 79%, higher than Escherichia coli (5% - 22%) (4). Klebsiella species are the third cause of ventilator-associated pneumonia among patients admitted to intensive care units (ICUs) and account for 83% of HAI cases (3, 5). Klebsiella pneumoniae is also well known for its high resistance to different antibiotic classes, especially beta-lactams. Carbapenemase-producing K. pneumoniae has been established as a major cause of HAIs worldwide, with severe clinical outcomes. Therefore, a more comprehensive understanding of its antimicrobial resistance (AMR) genetic determinants is critical to combat the spread of highly resistant K. pneumoniae in medical care centers and the associated morbidity and mortality (6).
Beta-lactamases are enzymes produced by bacteria that break down the beta-lactam ring in beta-lactam antibiotics, such as penicillins, cephalosporins, carbapenems, and monobactams (7). All four beta-lactamase enzyme classes (A-D) have been reported in association with K. pneumoniae (8). Gene blaCTX-M is a class A extended-spectrum beta-lactamase (ESBL) gene, which is carried on and transferred by integrons and plasmids. Therefore, it can spread from resistant to non-resistant strains during outbreaks, making it an important target for surveillance. Similarly, blaGES is present on mobile genetic elements (MGEs) and can induce partial resistance to carbapenems (9).
Metallo-beta-lactamase (MBL) genes have also been established as important pathogenesis factors, as their products confer significant resistance to most beta-lactam antibiotics (including carbapenems) and resistance to beta-lactamase inhibitors. One of these genes is blaIMP, which encodes a class B beta-lactamase (10). It is, therefore, essential to gain as much knowledge as possible about the prevalence of these genes to guide decisions about antibiotic administration and infection control efforts in healthcare settings, especially ICUs.
2. Objectives
This study aimed to investigate the AMR pattern of K. pneumoniae clinical isolates obtained from the sputum of patients hospitalized in ICUs of Rasool-e-Akram Hospital, Tehran, Iran, and to determine the presence of two ESBLs genes, i.e., blaCTX-M and blaGES, as well as the blaIMP metallo-beta-lactamase gene.
3. Methods
3.1. Clinical Samples, Bacterial Isolates, and Antibiotic Susceptibility Testing
Sputum samples were obtained from 105 patients hospitalized at ICU (1) respiratory ICU 1; and (2) emergency ICU, neurology ICU, cardiac care unit, medical ICU, and ICU 4 wards of Rasool-e-Akram General Hospital Complex in Tehran, Iran, during February 2020-February 2021. The samples were then immediately transferred to the research laboratory in a sterile container containing 10 mL of normal saline for further investigations. Klebsiella pneumoniae isolates were identified using standard microbiological and biochemical tests. All media used were purchased from Merck, Germany.
Identification was confirmed by species-specific 16S rRNA sequence amplification using the primers presented in Table 1. Briefly, 3 µL of each sample was mixed with 10 µL Taq PreMix, 1 µL water, and 1 µL of each forward and reverse primers. The polymerase chain reaction (PCR) reaction included an initial denaturation of 1 min at 94°C, 35 denaturation cycles of 1 min at 94°C, annealing for 1 min at 58°C, extension for 1 min at 72°C, and a final extension of 7 min at 72°C. All PCRs were conducted using an Eppendorf mastercycler gradient (Eppendorf AG, Germany).
Primers Sets Used to Detect the 16S rRNA, blaCTX-M, blaGES, and blaIMP Genes
Antibiotic susceptibility testing (AST) was performed for all K. pneumoniae isolates using the Kirby-Bauer disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) 2021 guidelines (15). All antibiotic disks were procured from Padtan Teb, Iran. The following antibiotic disks were used as described before (16): Piperacillin-tazobactam (100/10 μg), ampicilin-sulbactam (10/10 μg), ciprofloxacin (5 μg), cefotaxime (30 μg), ceftazidime (30 μg), gentamicin (10 μg), amikacin (30 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), imipenem (10 μg), meropenem (10 μg), and colistin (10 μg).
3.2. Phenotypic Identification of ESBLs
In order to determine phenotypic ESBL production, the double disk synergy test was used according to the CLSI 2021 guidelines (15). For this purpose, the cefotaxime disk (Padtan Teb, Iran) was placed at a distance of 20 - 30 mm from the cefotaxime/clavulanic acid disks. Similarly, the ceftazidime disk was placed at a distance of 20 - 30 mm from the ceftazidime/clavulanic acid disk. Clear areas (non-growth) between the two disks indicated ESBL production.
3.3. Molecular Identification of the 16S rRNA, bla<sub>CTX-M</sub>, bla<sub>GES</sub>, and bla<sub>IMP</sub> Genes
Polymerase chain reaction was performed using the primers provided in Table 1 and thermal cycling programs and PCR mixtures (2X Taq PreMix, Parstous, Iran) based on previously described methods (11-14) to detect the K. pneumoniae16S rRNA gene as well as the blaCTX-M, blaGES, and blaIMP genes. The PCR setting for each gene was as follows: (A) initial denaturation: 1 min at 94°C for all genes; (B) 35 cycles of denaturation (45 sec at 94°C, 30 sec at 95°C, and 30 sec at 95°C for CTX-M, GES, and IMP, respectively ), annealing (45 sec at 54°C, 30 sec at 60°C, and 30 sec at 50°C for CTX-M, GES, and IMP, respectively) and extension (1 min at 72°C for all genes); and (C) final extension: 10 min at 72°C for all genes. Polymerase chain reaction products were then run on a 1% agarose gel electrophoresis for confirmation.
3.4. Statistical Analysis
Data are demonstrated as mean ± standard deviation or number (percentage) based on the nature of the datum. All graphs used in this manuscript were created using the GraphPad Prism 9 software (GraphPad, USA).
4. Results
4.1. Clinical Samples and Bacterial Isolates
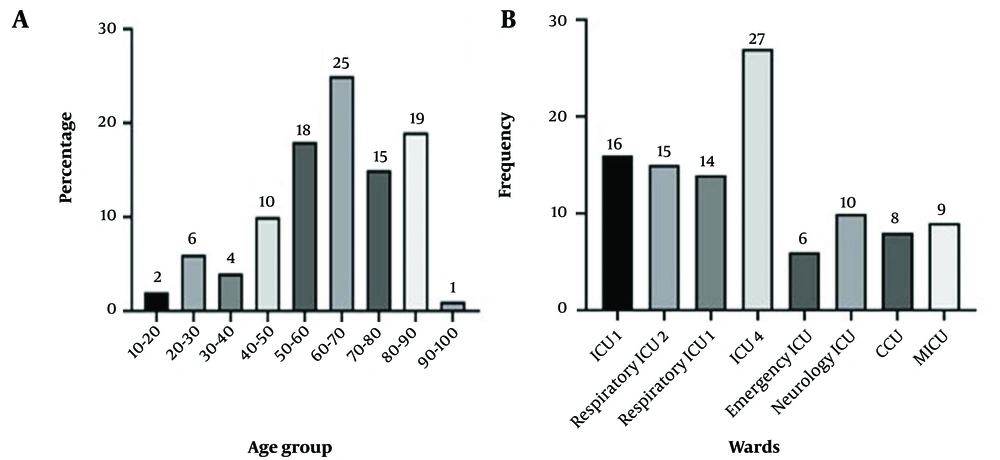
A total of 105 K. pneumoniae clinical isolates were collected from different ICUs, as demonstrated in Figure 1B. Of these, 58 (55.2%) studied patients were male and 47 (44.8%) were female. The mean age of the patients was 57.10 ± 20.03 years. About 19.04% and 17.14% of the patients were in the age ranges of 70 - 80 and 60 - 70 years, respectively (see Figure 1A). As demonstrated in Table 2, the most frequent comorbidities among patients were peripheral vascular disease (23/105, 21.9%), diabetes (22/105, 21%), and bone fracture (21/105, 20%).
| Values a | |
|---|---|
| Age | 57.10 ± 20.03 |
| Gender | |
| Male | 58 (55.2) |
| Female | 47 (44.8) |
| Comorbidities | |
| Diabetes | 22 (21) |
| Cancer | 12 (11.4) |
| Renal failure | 14 (13.3) |
| Lung disease | 3 (2.9) |
| Peripheral vascular disease | 23 (21.9) |
| Bone fracture | 21 (20) |
| Alzheimer | 10 (9.5) |
Demographic Information of the Study Population
4.2. Antimicrobial Resistance Patterns
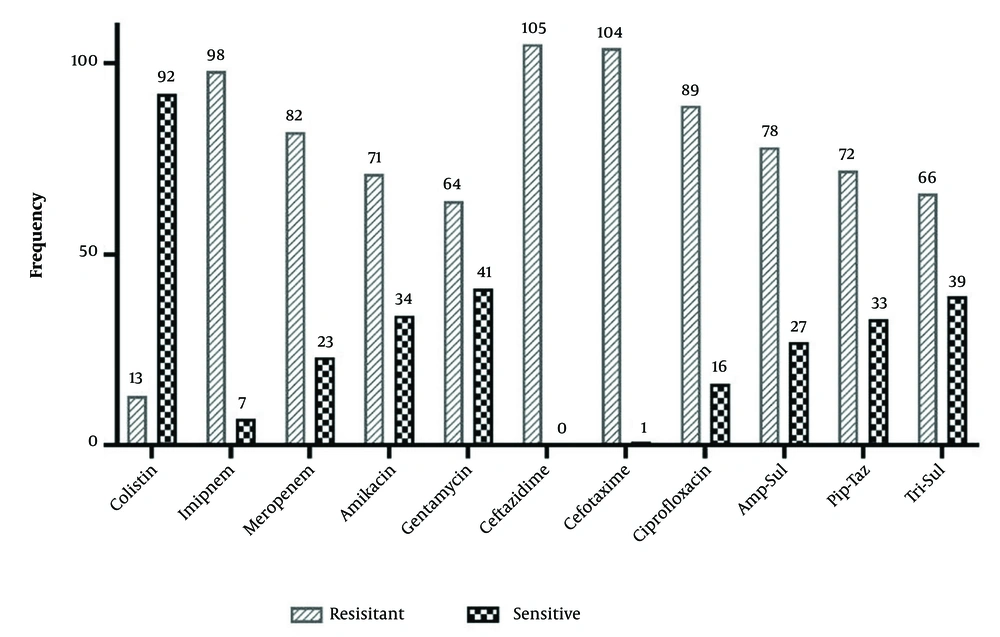
The results of AST for K. pneumoniae isolates are presented in Figure 2. The highest resistance was observed for ceftazidime (100%), cefotaxime (99%), and imipenem (93.3%), while resistance to colistin (12.4%) was the least. We observed that 16 isolates were phenotypically identified as ESBL producers.
4.3. Presence of bla<sub>CTX-M</sub>, bla<sub>GES</sub>, and bla<sub>IMP</sub>
Among 105 clinical isolates, 91 (86.7%) harbored the blaCTX-M ESBL gene (Figure 3). The blaGES and blaIMP genes were not detected in any of the isolates.
5. Discussion
It has been estimated that 17 million people die globally as a result of infectious diseases every year. According to a report by the United States Center for Disease Control, the number of annual deaths due to multidrug-resistant (MDR) infections will reach 10 million, and the mortality of bacterial infections will exceed heart diseases and cancer if no new treatments are developed. Infectious diseases are currently the second leading cause of death worldwide and the fourth leading in the US (17). Approximately 80% of enterobacterales-related carbapenem-resistant bacterial infections are caused by K. pneumoniae. In addition, K. pneumoniae causes approximately 12% of all hospital-acquired pneumonia cases (17).
The bacterial pneumonia caused by this bacterium is different from that caused by other agents, such as Streptococcus pneumoniae, in that patients with K. pneumoniae infections produce thick and yellow-brown sputum, which is indicative of extensive inflammation and necrosis in the respiratory tissue (17). This study determined the AMR pattern of K. pneumoniae clinical isolates, investigated the phenotypic production of ESBLs, and evaluated the frequency of the blaCTX-M, blaGES, and blaIMP genes.
All isolates were found to be resistant to at least three classes of antibiotics and were considered MDR. The highest resistance was observed against the third-generation cephalosporins, including ceftazidime (100%) and cefotaxime (99%). The resistance rate to carbapenems (imipenem 93.3% and meropenem 78.1%) and fluoroquinolones (ciprofloxacin 84.8%) was also very high. Similar to our study, Jalalvand et al. collected 800 enterobacterales clinical isolates from hospitals, of which 291 were K. pneumoniae and 66.66% were obtained from ICUs. In their study, 108 K. pneumoniae isolates were resistant to carbapenems and all cephalosporins, of which 102 were obtained from respiratory samples (18). These similar rates of antibiotic resistance may indicate the common sources of infection or resistance genes. Similar medical practices in terms of antibiotic administration may have also contributed to the development of similarly high antibiotic resistance rates.
In a study on the hospital and environmental isolates of K. pneumoniae in Mexico, Cordova-Espinoza et al. reported relatively lower rates of resistance to similar antibiotics, which could indicate higher AMR rates in ICUs compared to other hospital wards and the environment (19). This could be due to the closer proximity of patients or poorer infection control practices in ICUs compared to other hospital wards. In a systematic review of K. pneumoniae AMR in Asia, Effah et al. reported similar but marginally lower resistance rates for most tested antibiotics. However, our isolates showed notably higher resistance rates to colistin and carbapenems (20). This could indicate a more serious antibiotic resistance problem in Iran compared to other Asian countries, or it could be attributed to differences in sampling periods and sources.
Polymyxins, especially colistin, are among the few agents that retain their efficacy against carbapenem-resistant K. pneumoniae. However, the ever-increasing administration of these antibiotics has contributed to the emergence of colistin-resistant strains (21). Almost 12.4% of our isolates were resistant to colistin. Resistance to colistin has been increasingly reported from all parts of the world, including the Middle East region. A high resistance rate of 16.9% was reported during 2015 - 2016 from Iran (22). Research in Iran revealed an increase of up to 50% in colistin resistance in carbapenem-resistant K. pneumoniae isolates (23). Although colistin resistance has been reported in other countries, higher rates of resistance were observed in our isolates compared to them (24). As with other instances of increased antibiotic resistance, excessive antibiotic administration, poor infection control practices, and horizontal transfer of resistance genes are possible contributing factors. It is essential to design and implement standard practices for monitoring the use of antibiotics and controlling nosocomial infections to reduce the spread of antibiotic-resistant strains.
The frequency of the blaCTX-M gene in our isolates was 86.7%, which explains the high level of resistance to cefotaxime and ceftazidime antibiotics. Similarly, the prevalence of blaCTX-M was reported as high as 100% and 89.2% by Patil et al. and Rameshkumar et al., respectively (25, 26). An investigation in Iran in 2018 showed that 88 out of 94 K. pneumoniae isolates harbored blaTEM, blaSHV, and blaCTX-M-15 concurrently, which is in line with our results (27). These similar rates may indicate an exogenous source of resistance genes, as the frequencies of these genes are similar in different regions. No isolates were positive for blaGES in this study. In the research conducted by Patil et al., the prevalence of blaGES was 9%, while Indrajith et al. reported its prevalence as high as 20% in 2021 (25, 28).
Carbapenem-resistant K. pneumoniae has spread extensively in medical care settings. More than 90% of our isolates were carbapenem-resistant. In 2017, Moemen and Masallat collected 125 K. pneumoniae isolates, of which 42 were carbapenem-resistant, and 62% were recovered from respiratory specimens. In their study, the highest rate of resistance was reported against cefotaxime (100%) and ceftazidime (97.6%), similar to the present study. Moreover, resistance to carbapenems, including meropenem, imipenem, and ertapenem, was reported to be 71.4%, 59.5%, and 92.9%, respectively (29).
Resistance to carbapenems can be caused by the production of K. pneumoniae carbapenemase, New Delhi metallo-β-lactamase (NDM), MBLs, oxacillinase-48 (OXA-48), ESBLs, and porins as well as the hyperproduction of Ambler class C (AmpC) β-lactamase (30). The blaIMP gene was the only carbapenemase gene investigated in our study, which was not detected in any isolates. It is very likely that carbapenemase genes other than blaIMP were responsible for carbapenem resistance in our isolates (29). Ssekatawa et al. reported the presence of blaIMP as high as 19.4% in 2021 (31). Hu et al. in China collected 159 carbapenemase-producing K. pneumoniae isolates during 2018 - 2019, of which 50.9% were recovered from sputum samples.
All isolates were MDR and resistant to imipenem, meropenem, gentamicin, cefoxitin, ceftazidime, cefoperazone/sulbactam, and aztreonam. The prevalence of resistance to imipenem was reported to be more than 90%, and blaKPC was positive in 81.1% of the isolates. No blaIMP and blaGES were identified in their study, which is in line with our results (32). It could be construed that blaIMP as a resistance gene is not a point of concern currently in our region. In contrast, other beta-lactamase genes, such as blaCTX-M, are much more prominent and should receive higher priority in surveillance programs.
5.1. Conclusions
Periodic examination of the phenotypic and genotypic resistance patterns of patients is highly effective in combating AMR and leads to decreased hospitalization and medical care costs. Other ESBL genes can also be investigated for more precise prediction of AMR status in the clinical isolates of K. pneumonia.