1. Background
Candida albicans is an opportunistic fungal pathogen that colonizes various human body parts such as the oral cavity, throat, gastrointestinal tract, vagina, and skin in healthy individuals. It is a part of the body's normal flora. Under certain conditions, these opportunistic microorganisms can lead to infections (1, 2). Several virulence factors, including adhesin expression, hydrolytic enzyme production, the ability to switch from yeast to hyphae, and metabolic adaptability, enable C. albicans to proliferate and invade host tissues (3). The most recurrent and severe infections are often associated with the formation of biofilms on biological or artificial surfaces.
Biofilm formation by C. albicans significantly enhances its resistance to conventional antifungal treatments and the host's immune defenses, making infections difficult to eradicate (4). Key genes regulating biofilm formation include BCR1, ALS1, ALS3, HWP1, and ECE1, which are considered promising therapeutic targets. Previous research has shown that specific compounds can inhibit biofilm formation by suppressing these genes (5). Streptomyces cellulosae, a species of Actinobacteria known for producing bioactive compounds with antimicrobial properties, has shown potential against various pathogens, including fungi and harmful Gram-positive bacteria (1, 2). Notably, Mahmood et al. found that S. cellulosae produces compounds that inhibit biofilm formation by Pseudomonas aeruginosa (6).
2. Objectives
Given the rising number of infections and concerns about resistance, it is crucial to develop new strategies to combat biofilm-forming organisms like C. albicans. Therefore, this study aimed to evaluate the effectiveness of extracts from S. cellulosae in targeting genes associated with biofilm formation in C. albicans.
3. Methods
3.1. Isolation and Identification of Microorganisms
Pure cultures of S. cellulosae were sourced from the Charmo Research Center at Charmo University, Sulaimaniyah, Iraq. The bacterium was cultivated on Gause's medium after being extracted from soil and identified using colony morphology and 16S rDNA genomic sequencing. Sequence analysis confirmed that the bacteria matched those in the NBRC 13027 strain (6). Clinical samples for isolating C. albicans were collected from Hiwa Hematology/Oncology Hospital in Sulaymaniyah, Iraq. Samples were taken using swabs (7) from symptomatic leukemic patients' oral lesions, then inoculated on Sabouraud dextrose agar (SDA) and incubated at 37°C for 24 hours (8). Isolates were further cultivated on CHROM agar Candida and incubated at 37°C for 48 - 72 hours (9). Colony morphology, including color, size, and texture, was examined to identify the species (10).
3.2. Antibiofilm Formation by Streptomyces cellulosae
Initially, S. cellulosae was cultured using the batch culture method (11), and 100 µL of bacterial spore stock (1×108 spores/mL) was added to 100 mL of tryptic soy broth (TSB) (12). The cultures were maintained in a shaking incubator (LSI-5004M, Indonesia) at 30°C for 48 hours at 150 rpm. Subsequently, TSB was inoculated with a 5% v/v inoculum and cultured for 120 hours at 37°C (13). The bioactive compounds from the broth culture were then extracted. The culture filtrate was mixed with ethyl acetate (1:1, v/v) and vigorously shaken for 20 minutes. Using a rotary evaporator (Heidolph, GmbH, Germany), the ethyl acetate layer was separated from the liquid phase and concentrated by evaporation at 40°C until dry. The resultant residue was dissolved in pure methanol to produce a reddish-brown crude extract and stored at -20° C until needed.
3.3. Biofilm Formation by Candida albicans
Biofilm development was evaluated using 96-well microplates (14). Briefly, C. albicans isolates (McFarland 1 × 108 CFU/mL) were cultured overnight in SDA broth at 37°C (12). After incubation for 24 hours at 37°C in 2.0 mL of yeast extract peptone dextrose (YPD) broth, tubes were diluted (1:20) with YPD supplemented with 1% glucose and incubated for another 24 hours at 37°C (15). The optical density (OD) was then measured at 630 nm. Each test was conducted in triplicate. Subsequently, the OD of each strain (ODs) was compared to that of the negative control (ODnc). The reference strain of C. albicans (ATCC 90028) served as the positive control for biofilm formation, whereas the negative control comprised solely of broth media without C. albicans. Strains were categorized based on their biofilm-forming ability as none (ODs = ODnc), weak (ODnc < ODs < 2ODnc), moderate (2ODnc < ODs < 4ODnc), and strong (4ODnc < ODs) (15).
3.4. Antibiofilm Activity of Streptomyces cellulosaeagainst Candida albicans
The antibiofilm efficacy of Streptomyces isolates against C. albicans was determined using a crystal violet assay (16, 17). The previously formed C. albicans biofilms in 96-well microplates were treated with 0.5, 1, 2, 3, and 4 µg/mL of the S. cellulosae antibiofilm extract (100 µL) for 48 hours at 37°C. Subsequently, the medium was discarded, and the wells were washed with sterile phosphate-buffered saline (PBS), inverted for blotting, and air-dried. Each well then received 200 µL of methanol for fixation and was incubated for 15 minutes at 25°C. After discarding the methanol, the plates were air-dried for 45 minutes. Next, 200 µL of a 0.1% w/v crystal violet solution was added to each well, and the plate was incubated for 20 minutes at 25°C. Following two washes with distilled water, 200 µL of an acetone: Ethanol (20:80 v/v) mixture was added to each well and incubated for 15 minutes at 25°C to stain the biofilm (18). Biofilm production was quantified using a plate reader by transferring 100 µL from each well to a new, sterile microplate and measuring the OD at 630 nm. Each test was repeated three times (15).
3.5. Quantitative Real-time PCR Analysis of Candida albicans Biofilm-Specific Genes
A 96-well polystyrene microtiter plate (Iwaki), flat-bottomed and pre-sterilized, was used to culture a standard cell culture of C. albicans (1.0 mL). After incubating with agitation at 37°C for 1.5 hours, the liquid was discarded and the wells were rinsed twice with PBS. Each well was then replenished with 1.0 mL of fresh YPD medium containing 3.0 mg/mL of Streptomyces crude extract and incubated for 24 hours at 37°C. Post-incubation, the supernatant was removed and the wells were washed twice with PBS. Total RNA from C. albicans biofilms was isolated using the SV Total RNA Isolation System (Biolab), and 2.0 mg of RNA was reverse-transcribed using Superscript II (Invitrogen). The resulting cDNAs were PCR-amplified, with sequencing confirming product specificity. quantitative real-time PCR (qRT-PCR) primers were designed specifically for the C. albicans biofilm-associated genes ALS1, ALS3, BRG1, EFG1, HSP9, HWP1, and ACT1 (Table 1).
| Genes | Sequence | Reference |
|---|---|---|
| ALS1 | F: GACTAGTGAACCAACAAATACCAGA | (19) |
| R: CCAGAAGAAACAGCAGGTGA | ||
| ALS3 | F: CCAAGTGTTCCAACAACTGAA | (20) |
| R: GAACCGGTTGTTGCTATGGT | ||
| EFG1 | F: TATGCCCCAGCAAACAACTG | (21) |
| R: TTGTTGTCCTGCTGTCTGTC | ||
| hsp90 | F: GCTTTAAGTGCTGGTGCTGACGTT | (22) |
| R: TGGTACCACGACCCAATCTTTCGT | ||
| HWP1 | F: GACCGTCTACCTGTGGGACAGT | (23) |
| R: GCTCAACTTATTGCTATCGCTTATTACA | ||
| BGR1 | F: ACGATCAACCATTAGTGGAGG | (24) |
| R: GAAGAAGTAGGTGTAGATGATCCAC | ||
| ACT1 | F: TTTCATCTTCTGTATCAGAGGAACTTATTT | (25) |
| R: ATGGGATGAATCATCAAACAAGAG |
The RT-PCR mixture (20 µL) included 10 µL of Luna Probe One-Step Reaction mix (No ROX) (2 ×), 1.0 µL of Luna WarmStart RT Enzyme Mix (20 m ×), 0.8 µL of primers, 0.4 µL of Probe (10 µL), template RNA, and 20 µL of nuclease-free water. The Roche LightCycler® was utilized for qRT-PCR, with each run comprising a 10-minute pre-incubation at 95°C, followed by 40 cycles of 10 seconds of denaturation at 95°C and 30 seconds of annealing/extension at 60°C. ACT1 served as the reference housekeeping gene. Each concentration of crude extract was tested in at least three experimental repeats.
3.6. Growth Rate Measurements
Candida albicans cells were cultivated overnight in SDA at 37°C with shaking at 200 rpm. After obtaining two cell counts, 20 mL of fresh SDA was inoculated with the cells at a density of 1 × 106 cells/mL. Spectrophotometric readings were taken hourly, with the cultures shaken at 200 rpm and maintained at 37°C. Growth rates were monitored over three days for various initial cultures.
3.7. Statistical Analysis
Statistical analyses of the qRT-PCR results were performed using Microsoft Excel and Graph Pad PRISM 8. The average Ct values for the genes ALS1, ALS3, BRG1, EFG1, HSP9, HWP1, and ACT1 were presented as mean ± standard deviation (SD) before and after treatment with the crude extract. A P-value of < 0.05 was considered statistically significant.
4. Results
4.1. Antibiofilm Activity of Streptomyces cellulosae Crude Extract
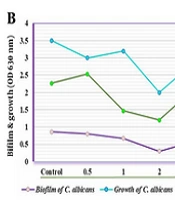
The study demonstrated that the crude extract from S. cellulosae inhibited biofilm formation by C. albicans at various concentrations (0.5, 1.0, 2.0, 3.0, and 4.0 µg/mL).
4.2. Gene Regulation of Biofilm Formation in Candida albicans
The efficacy of S. cellulosae crude extract against C. albicans showed that concentrations above 0.5 µg/mL notably impeded biofilm formation by the Candida isolates. Additionally, dosages greater than 2.0 µg/mL significantly inhibited Candida biofilm production (P < 0.05).
4.3. Antibiofilm Activity of Streptomyces cellulosae Crude Extract on the Expression of Biofilm-Related Genes in Candida albicans
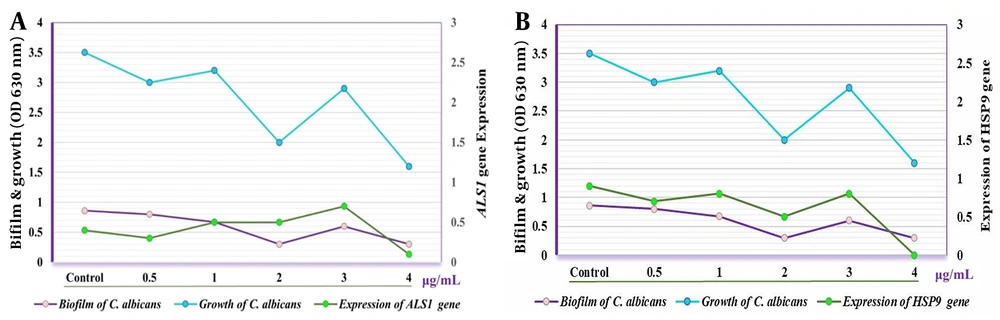
Quantitative real-time PCR analysis revealed the impact of the crude extract on the expression levels of biofilm-associated genes in C. albicans at concentrations ranging from 0.5 to 4.0 µg/mL. Gene expression was normalized against ACT1, used as the reference gene. The expression of ALS1 and ALS3 genes decreased significantly at concentrations of 0.5 and 4.0 µg/mL, and at 2.0 and 4.0 µg/mL respectively, compared to the control group (CG) which received no treatment (P < 0.02). Additionally, the expression levels of ALS1 and ALS3 increased significantly at a concentration of 3.0 µg/mL compared to CG (P < 0.03) (Figure 1).
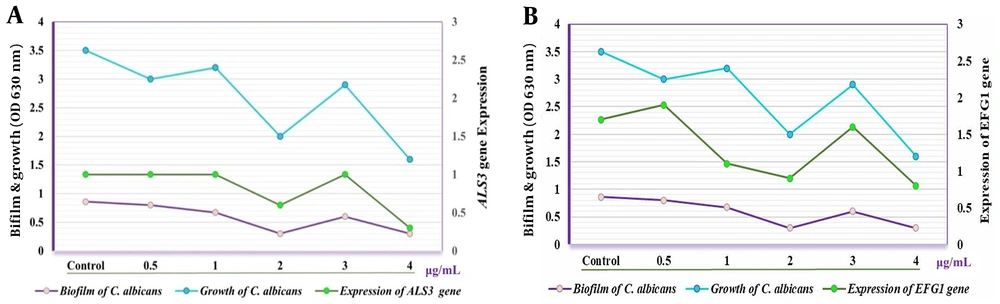
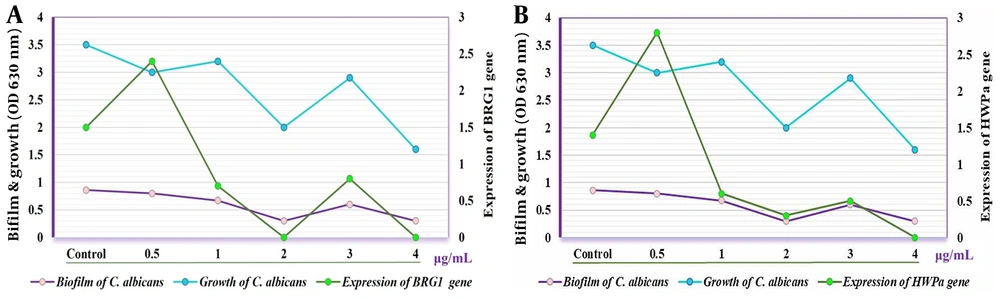
Furthermore, the expression of EFG1 was markedly higher after treatment with the crude extract at 0.5 and 3.0 µg/mL compared to CG (P < 0.05) and decreased progressively at concentrations of 1.0, 2.0, and 4.0 µg/mL. Conversely, the HSP9 gene maintained normal expression levels following treatment with the crude extract (Figure 2). The expression of both HWPa and BRG1 genes was notably affected at concentrations of 0.5 and 3.0 µg/mL. Notably, a significant reduction in the expression of HWPa and BRG1 genes was observed at concentrations of 2.0 and 4.0 µg/mL when the C. albicans isolate was treated with the crude extract (Figure 3).
5. Discussion
Candida is the fourth most common cause of hospital-acquired infections (26) and a leading contributor to biofilm formation associated with medical interventions, drug use, and implants (27). Consequently, this study focused on the local impact of these biofilms. Our findings showed that concentrations of the crude extract above 0.5 µg/mL significantly inhibited Candida biofilm formation, with even more pronounced effects observed at concentrations exceeding 2.0 µg/mL. Furthermore, the study revealed that the expression levels of the ALS1 and ALS3 genes varied under different concentrations of the crude Streptomyces extract, generally decreasing at most concentrations but increasing at 3.0 µg/mL. Research by Deng et al. highlighted the influence of ALS3 gene expression on the biofilm formation capabilities of C. albicans, suggesting that manipulating this gene's expression could aid in developing both therapeutic and preventive strategies (28), a finding that corroborates our results. Additionally, Martorano-Fernandes et al. demonstrated that gene expression levels, including those of the ALS1, ALS3, and HWPa genes, could vary based on the environmental conditions and substances present, noting that increased gene expression could exacerbate pathogenic conditions (29).
Zarimeidani et al. explored the minimum inhibitory concentrations (MIC) of caprylic acid/nanoencapsulated caprylic acid against C. albicans, ranging from 625 - 400 to 1.3-50 µL/mL. Their study utilized qRT-PCR to measure the expression level of the EFG1 gene, finding the highest activity of caprylic acid at 450 and 500 µg/mL, with the most potent activity for nanoencapsulated versions at 6.2 and 3.1 µg/mL (30). Another innovative approach involves combining inhibitors of Histone Deacetylase (HDAC) and Heat Shock Protein 90 (HSP90) to enhance the efficacy of fluconazole against resistant strains of C. albicans. This pharmacological strategy was shown to reduce gene expression related to drug resistance and mitigate major pathogenic factors, presenting a contrast to our findings where gene expression varied based on treatment concentration and gene targeted (31). These differences could stem from variations in experimental methods and the specific genes analyzed.
Antimicrobial peptides (AMPs) have emerged as a promising approach to treat Candida infections. Samot and Rouabhia investigated the antimicrobial effects of Dermaseptin S4 (DS4) against C. albicans, focusing on its impact on the growth, morphological alterations, biofilm formation, and gene expression of the fungus. DS4 halted the growth of C. albicans at concentrations of 32 and < 16 µg/mL. It appears to obstruct the yeast-to-hyphae transition and limit biofilm development by reducing biofilm mass weight. Significantly, Dermaseptin also reduced the expression of the HWP1 and EAP1 genes (32). The polymorphic nature of C. albicans, which can grow in both yeast-like and filamentous forms, makes the transition between these states crucial for its pathogenicity. EFG1 and BRG1 are important transcriptional regulators that influence the growth and morphological transformations of this fungus, impacting its pathogenicity and biofilm formation (33-35). Biofilms are microbial communities that form structures by attaching to surfaces, and can include combinations of bacterial or fungal species (36). The inhibitory effect of S. cellulosae on biofilm synthesis was also observed in Iraq by Mahmood et al., supporting the current investigation that demonstrates S. cellulosae's ability to produce metabolites with antibacterial and antibiofilm properties (6)
5.1. Conclusions
It is concluded that the crude extract of S. cellulose has potential as a novel antibiofilm agent against C. albicans by suppressing the expression of some involved genes, offering a promising avenue for future research and potential therapeutic applications.