1. Background
Hepatitis B virus (HBV) is a major etiological agent of hepatitis globally (1), with chronic cases leading to end-stage liver diseases such as hepatocellular carcinoma (HCC) and cirrhosis (2). The prevalence of HBV infection among the Iranian population is approximately 3%, and the progression of related liver disease is influenced by host, viral, and environmental factors (3). Mutations in the viral genome play a critical role in exacerbating the infection (4). Some genomic regions, such as the pre-core/basal core promoter (BCP), direct repeat sequences, and enhancer II (EnH II), overlap with the HBV X-gene (5). The HBX protein, an important non-structural protein with transcriptional transactivator capabilities, affects both cellular and viral promoters (6). It functions as a multifaceted oncoprotein related to HCC in chronic HBV patients (CHB) through a multistep process, which includes the induction of reactive oxygen species, alteration of mitochondrial function and physiology, and DNA damage (7, 8).
Different genetic variations in the X-gene, such as mutations during chronic infection (9), can impact not only the amino acid sequence but also HBV expression and the expression of other genes (10). These mutations can influence the physiological functions of the HBX protein, affecting biological mechanisms such as cell proliferation, transcription, apoptosis, and signal transduction, ultimately contributing to disease progression (11). The HBX protein is also able to create a cellular environment conducive to HBV replication by activating host genes associated with cell proliferation and inflammation (12).
In hepatic inflammation, HBX stimulates the transcription of numerous pro-inflammatory cytokines. The protease inhibitor-like structure of HBX may also cause the accumulation of toxic factors, resulting in severe hepatocellular injury (8, 13). Specific mutations such as AG1762/1764TA, C1653T, C1485T, T1753C, A1383C, and G1613A have been associated with HCC survival (14). The nonsynonymous C1653T mutation in the EnH II region may alter binding affinity, leading to an amino acid substitution in the HBX gene (15). The occurrence of the double mutation K130M + V131I increases as liver disease progresses and may contribute to HCC by affecting NF-κB activity (16, 17).
2. Objectives
Recent studies have reported that HBX plays a crucial role in the progression and pathogenesis of HBV-related complications. Therefore, the aim of this study, similar to our previous research on S-gene mutations in HBV, was to analyze and detect X-gene mutational patterns in patients across three generations.
3. Methods
3.1. Baseline Demographic Features of the Study Population
In this cross-sectional study conducted in the northeastern region of Iran, from September 2020 to January 2021, at the Research Center of Gastroenterology and Hepatology, Golestan University of Medical Sciences, 3,250 HBV-infected patients were investigated based on the inclusion criteria. Samples were collected from patients with CHB, and plasma was separated from the blood and stored. All participants were tested for alanine transaminase (ALT), aspartate transaminase (AST), and alkaline phosphatase (ALP) levels (18). Ninety patients with confirmed HBV infection (positive HBs-Ag, history of HBV infection for more than six months) were included in the study. Patients were divided into three groups: Three-generations group (30 cases), two-generations group (40 cases), and intrafamilial group (20 cases), based on the inclusion criteria (HBs-Ag: Positive, HBs-Ab: Negative, HBc-Ab: Positive, HBe-Ag and HBe-Ab: Positive/negative). Informed consent was obtained from each participant after a thorough interview explaining the study goals and answering any questions. Participation in the study was voluntary and had no impact on the treatment course of patients.
3.2. Extraction of Hepatitis B Virus DNA and Semi-Nested PCR
The HBV-DNA extraction was performed from 200 μL of serum following the manufacturer's instructions (Viral Nucleic Acid Extraction Kit, Yektatajhiz). Amplification of HBV sequences covering nucleotides 1365 - 2078 was conducted using F1/R1 primers. In the second round, a 713 bp template was used to amplify nucleotides 1365 - 1881 with F1/R2 primers (Table 1) (4).
| Primer Name | Sequence (5' to 3') | Target Sequence | PCR Program |
|---|---|---|---|
| F1 | ATCGTATCCATGGCTGCTAGGCT | 1365 - 1387 | Step 1: 94°C 5 min, 35 cycles (94°C 1 min, 55°C 1 min and 72°C 1 min), 72°C 7 min; Step 2: 94°C 5 min, 35 cycles (94°C 30 Sec, 57.5°C 30 Sec and 72°C 45 Sec), 72°C 5 min. |
| R1 | CAGAATAGCTTGCCTGAGTGC | 2058 - 2078 | |
| R2 | CACAGCTTGGAGGCTTGAACA | 1861 - 1881 |
3.3. DNA Sequencing and Mutation Analysis
PCR products that yielded positive results were subjected to automated directional sequencing (3130 Genetic Analyzer, ABI/HITACHI, Tehran University of Medical Sciences). Mutation identification and analysis were performed by aligning the nucleotide sequences with the standard sequence of hepatitis B (accession number: AB033559) from the GenBank database. Statistical analysis was conducted using SPSS version 20. The student’s t-test was used to compare the collected data, with a P-value of less than 0.05 considered statistically significant for differences between categories.
4. Results
Investigation of the HBx gene through sequencing of 90 samples from patients (accession numbers: ON346437-ON346526) was conducted. In contrast to the other groups, liver function test results in the three-generation group showed a significant association with values above the normal range. The study found that in the intra-familial group and the two-generation group, liver function test levels were elevated in HBe-Ag-positive patients, with statistical significance observed in the two-generation group (P-value = 0.08). In the third group, there was no significant correlation between liver function test levels, gender (P-value = 0.5), and age (P-value = 0.06) (Table 2).
| Basic Characteristics | Three Generations; (Grandmother- Mother and Child) | Two Generations; (Mother and Child) | Intrafamilial Members |
|---|---|---|---|
| Age | 45.23 ± 22.8 | 35.83 ± 17.7 | 35.5 ± 15.2 |
| Gender | |||
| Male | 4 (13.4) | 11 (27.5) | 8 (40) |
| Female | 26 (86.6) | 29 (72.5) | 12 (60) |
| Total | 30 | 40 | 20 |
| HBe-Ag | |||
| + | 0 (0) | 7 (17.5) | 13 (65) |
| - | 30 (100) | 33 (82.5) | 7 (35) |
| Total | 30 | 40 | 20 |
| Anti-HBe | |||
| + | 30 (100) | 4 (10) | 16 (80) |
| - | 0 (0) | 36 (90) | 4 (20) |
| Total | 30 | 40 | 20 |
| ALT (IU/L) | 24.6 ± 14.37 | 24 ± 8.9 | 22 ± 7.9 |
| AST (IU/L) | 25.46 ± 13.67 | 26 ± 11.2 | 23.3 ± 10.2 |
| ALP (IU/L) | 419.567 ± 259.78 | 464.75 ± 274.1 | 337.6 ± 172.2 |
Abbreviations: IU/L, international unit/litre; ALT, alanine transaminase; AST, aspartate transaminase; ALP, alkaline phosphatase.
a Values are expressed as mean ± SD or No. (%).
4.1. HBx Mutations Concerning the Status of Hepatitis B Virus Infection
When the sequences of the HBV partial genome isolated from CHB patients were compared with the reference sequence, it was found that 9.47% of CHB patients did not have a mutation in the X-gene (3.4% in the first group, 15% in the second group, and 10% in the third group). The sequence analysis of the X-gene in these patients demonstrated the highest degree of homology (79.3%) and the lowest mutation rate (20.7%) in mothers and children, respectively. Additionally, the results showed that grandmothers in the three-generation group had the most mutations in the X-gene region, which was statistically significant (P-value = 0.06).
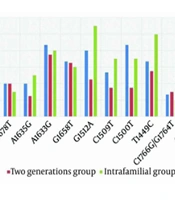
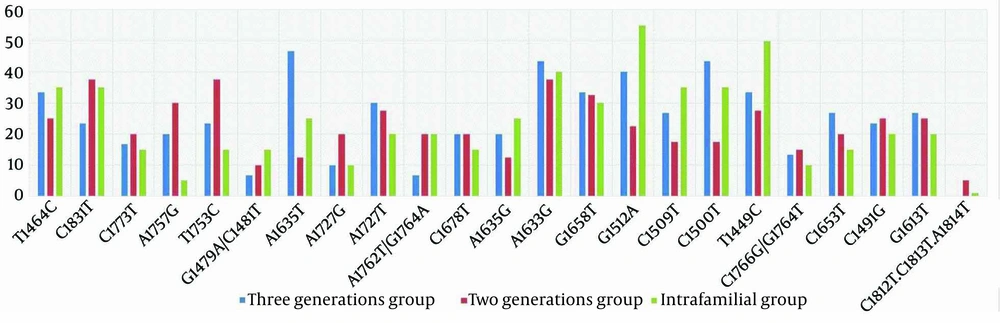
This study identified the co-occurrence of A1762T/G1764A mutations in 6.7% of the three-generation group, 20% of the two-generation group, and 20% of the intra-familial group. Statistical analysis revealed a significant difference (P-value = 0.03) in the co-occurrence of G1764A/A1762T mutations among these distinct groups. Co-occurrence of C1766G/G1764T mutations was observed in 13.4%, 15%, and 10% of CHB patients in the three groups, respectively (P-value = 0.4). The presence of T1464C was detected in 30% of CHB patients, with a higher prevalence in the intra-familial group (35%) compared to the two-generation (25%) and three-generation groups (33.4%) (P-value = 0.02). Additionally, the double mutation G1479A/C1481T was found in 6.7%, 10%, and 15% of CHB patients in the three groups, respectively (P-value = 0.04).
The C1500T point mutation was detected in 40% of CHB patients, with the highest frequency observed in the three-generation group (43.4%) compared to the other two groups (P-value = 0.03). The three-generation group also exhibited significantly higher frequencies of A1635T (46.7%) and A1635G (20%) point mutations compared to the other two groups (P-value = 0.01 and P-value = 0.04, respectively). Statistical analysis did not reveal a significant relationship between HBx mutations and elevated LFT or age (P-value = 0.1) (Figure 1).
The Hepatitis B virus DNA sequence analysis of the three-generation and two-generation groups revealed silent mutations A1727G (10% and 20%, respectively) and missense mutation A1727T (30% and 27.5%, respectively) (P-value = 0.04, P-value = 0.03, respectively). Additionally, a triplet mutation, A1762T/G1764A/C1773T, was detected in CHB patients. This study also identified another triplet mutation, specifically C1812T/C1813T/A1814T, which showed lower prevalence at 5% and 1% in the two-generation and intra-familial groups, respectively. However, no statistically significant difference was observed (P-value = 0.3) in the frequency of these mutations among the groups.
The B-cell epitope of the HBx protein showed two types of mutations: C1491G and C1500T. In the two- and three-generation groups, the prevalence of these mutations was 25% and 43.4%, respectively. The three-generation group also exhibited the highest abundance of mutations overall (33.4%) compared to the other groups. The G1613T mutation, which causes the substitution of Glu80 with Asp, was reported in 23.9% of the study population. Despite the comparable mutation frequency in both groups, statistical analysis did not reveal any significant difference (P-value = 0.2). According to our findings, the A1633G (43.4%), C1500T (43.4%), and A1635T (46.7%) mutations were the most abundant in the three-generation group compared to the other two groups.
4.2. Mutations in X Region and its Effect on HBe-Ag Expression
There was a statistically significant difference between CHB patients who tested positive for HBe-Ag and those who tested negative (P-value = 0.03). Among the 90 patients, only 11 (12.3%) tested positive for HBe-Ag. All CHB patients in the three-generation cohort tested negative for HBe-Ag. Additionally, it was observed that these patients had the highest number of mutations in the X region of the HBV genome.
5. Discussion
Detection of HBx in patients with HCC is a well-documented phenomenon often linked to mutations that contribute to HBV infection development (19, 20). HBx proteins lack the C domain, which is crucial for their suppressive effects on cell proliferation, growth, transactivation activity, and transformation (21, 22). In 2.9% of CHB cases, 8bp deletions or insertions in the C-terminus of HBx were found in the cirrhotic group. Moreover, 15 different deletions, such as 1769 - 1773, 1762 - 1768, 1763 - 1770, and T1771/A1775, were identified in cirrhotic patients (23). In contrast to previous research, this study found that CHB patients did not demonstrate any deletion-related mutations, except for C1773T, which had a 20% prevalence in the two-generation group. Our results align with previous studies (23, 24), showing that HBe-Ag-negative patients are more likely to have deletion and insertion mutations in the C-terminus region of HBx.
Another study demonstrated that the A1762T/G1764A mutation in the C-terminal overlap region of the X-gene with BCP contributes to the progression of the disease from the chronic phase to cirrhosis. Salarnia et al. showed that the A1762T/G1764A mutation was more common in patients with cirrhosis than in those with CHB (4). The presence of this mutation plays a significant role in the advancement of liver disease to more critical stages (4). Our findings are consistent with those of previous studies, including Chen et al. (16), Vazjalali et al., and Maleki et al. (25, 26), which showed that an increased prevalence of A1762T/G1764A mutations contributes to disease progression. This mutation accelerates viral replication, and our results indicate it is more frequent in HBe-Ag-negative patients in the two-generation group. We also found that the C1773T point mutation coincided with the A1762T/G1764A double mutation, though neither was statistically significant.
A study by Salarnia et al. identified new mutations within the HBx gene. Various mutations, including C1500T, C1491G, G1658T, and G1613T, were observed in the N-terminal region, Box α, Core promoter, and Enhancer II (6). Consistent with these findings, novel mutations were detected in CHB patients across the three-generation, two-generation, and intra-familial groups. The three-generation group exhibited the highest mutation frequency, with A1635T (46.7%), A1633G (43.4%), C1500T (43.4%), and C1491G (23.45%) being the most commonly reported. The occurrence of the A1635T mutation in the HBx protein sequence results in interaction with the DNA damage-binding complex-1 due to the overlap between the NRE and HBx coding sequences. A study by Ghosh et al. demonstrated a higher prevalence of A1635T among patients with cirrhosis (53.85%) compared to inactive HBV carriers and those with chronic HBV (27). In contrast to our results, this mutation was observed in CHB patients in the three-generation group.
Several studies have demonstrated that the A1727T mutation is a novel predictive marker for the cirrhosis phase in HBV-infected patients (6, 28). Patients with cirrhosis show a higher occurrence of the A1727G mutation, which increases the risk of HCC (29). The majority of TA1 mutations occur in the 1750-1755 nt region, which is recognized as a potential prognosticator of HCC (30). Additionally, the T1753C mutation has been identified as an important marker for cirrhosis severity and is associated with advanced liver disease (31). Consistent with these studies, we found that the A1727G mutation was present in 14.5% of CHB cases, with a statistically significant difference among the three groups (P-value = 0.07). The T1753C mutation, a key prognosticator of cirrhosis, was found in 23.4% of the three-generation group, 37.5% of the two-generation group, and 15% of the intra-family group.
It has been observed that specific patterns of HBx mutations can serve as early markers for an increased risk of HCC and predict clinical outcomes in HBV-infected patients. Prior research, such as that by Xiao et al., demonstrated that mutations in the X-region tend to emerge during the advanced phases of chronic HBV infection, leading to serious liver disorders such as HCC and cirrhosis (32). Our findings indicate that CHB patients exhibit significant occurrences of the double mutations C1481T and G1479A, as well as the point mutations A1635T, T1464C, and C1500T. Triple mutations, specifically A1762T/G1764A/C1773T and C1812/C1813T/A1814T, were observed in the two-generation and three-generation groups.
Furthermore, the absence of statistically significant differences in age and elevated LFT levels among individuals with HBx mutations is consistent with previous studies. The C1481T/G1479A mutations were found in all three patient categories, with a higher frequency observed in the two-generation and intra-familial groups compared to the three-generation group. Additionally, these mutations were more frequent in mothers than in their children and grandmothers, with this difference approaching statistical significance (P-value = 0.06). Our ability to comprehensively investigate disease progression and potential associations with combinations of HBV mutations is limited by the relatively small sample size of cases with progressive liver disease. We acknowledge that our findings may not be generalizable to other populations, particularly those infected with different HBV genotypes.
5.1. Conclusions
In conclusion, identifying and analyzing viral genomic mutations in correlation with clinical complications is crucial for disease management, prognosis, and treatment. These mutations can be used to screen high-risk individuals for liver disease, improve diagnostic methods, and refine therapeutic approaches. Additionally, further investigation into the impact of A1762T/G1764A mutations on different phases of HBV infection is recommended.