1. Background
The SARS-CoV-2 outbreak has emphasized the need for effective treatment and control strategies. Traditional research models face limitations, such as the lack of physiological relevance in 2D cultured cell lines (1, 2) and discrepancies in animal models (3). Understanding the infection mechanisms of SARS-CoV-2, particularly in the nasal cavity—the primary target for respiratory viruses—is crucial (4). However, limited information is available regarding its infection routes and replication in the nasal mucosal epithelium, necessitating a model that accurately reflects human nasal cavity infection to advance early detection and antiviral evaluations (5).
Organoids, three-dimensional cultures derived from stem cells, mimic the structures and functions of human organs (6-8). They are valuable tools for studying tissue biology, disease modeling, and drug discovery (9-11). While research on respiratory organoids in the context of SARS-CoV-2 infection exists (12-15), studies on nasal mucosal organoids (NMOs) are scarce. This research focuses on NMOs, cultivated from human nasal mucosal basal cells, which include ciliated and secretory cells (16). Nasal mucosal organoids exhibit characteristics of typical nasal epithelium and express ACE2 and TMPRSS2, both essential for SARS-CoV-2 infection.
2. Objectives
This study aims to develop an in vitro viral infection model using NMOs to accurately simulate human nasal mucosa. This model, combined with pseudo-viral tools, offers a more precise representation of the infection process than traditional models. By evaluating the efficacy of antiviral drugs, we aim to support the development of effective nasal treatments for SARS-CoV-2, contributing to epidemic prevention and clinical care.
3. Methods
3.1. Tissue Samples
Human NMOs were generated from tissues obtained from Nanfang Hospital, in compliance with ethical guidelines.
3.2. Viruses
The study utilized several SARS-CoV-2 virus and pseudovirus strains (Appendix 1), with all experiments conducted in a biosafety level 3 (BSL-3) laboratory.
3.3. Nasal Mucosal Organoid Culture
Nasal tissues were processed according to the method described for nasal mucosal tissue preparation (6), digested with enzymes (collagenase IV and dispase), and cultured in Matrigel within trans-well plates (Corning, USA). The culture medium (Accuroid, China) was replaced every 2 - 3 days.
3.4. SARS-CoV-2 Infection
Nasal mucosal organoids were seeded at a density of 2.0 × 104 cells per well in 100 μL and incubated at 37°C for 3 days. After the addition of the virus inoculum for 2 hours, the inoculum was replaced with culture medium for an additional 24 hours. Infection was assessed using fluorescence microscopy and a luciferase assay.
3.5. qRT-PCR
Total RNA was extracted from the organoids using TRIzol. The RNA was then converted to cDNA, and qRT-PCR was performed using custom primers (Appendix 2).
3.6. Flow Cytometry
Flow cytometry of NMOs was performed using an Attune CytPix flow cytometer (ThermoFisher, USA) with various cell marker antibodies detailed in Appendix 3.
3.7. Immunofluorescence
Post-infection, organoid samples were fixed, embedded, and processed for immunofluorescence analysis using primary and secondary antibodies to identify viral proteins.
3.8. Drug Treatment
Organoids were treated with camostat, remdesivir, and bergamottin (Appendix 4) for 1 hour prior to viral infection. Viral RNA was then measured by qPCR at 24 hours post-infection. To evaluate the impact of bergamottin, a CellTiter-Glo (Promega, USA) cell viability assay was conducted.
3.9. Statistics
Data were analyzed using three or more replicates, presented as mean ± standard deviation, and statistical significance was assessed using t-tests with a threshold of P < 0.05.
4. Results
4.1. Long-Term Culture and Validation of Nasal Mucosal Organoids
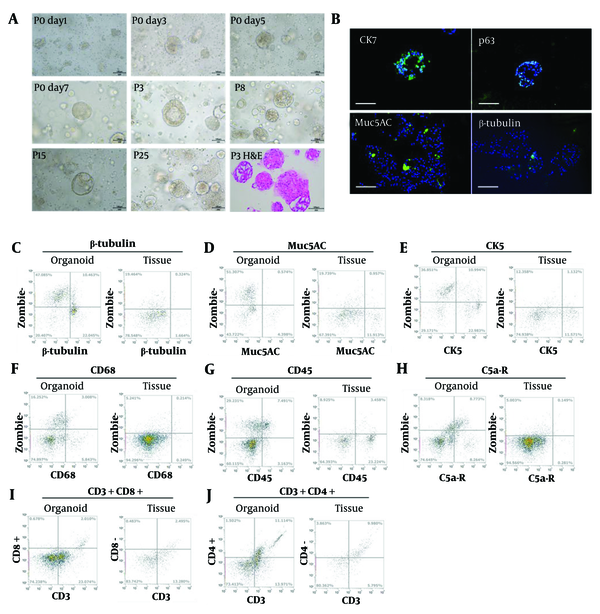
After seeding in Matrigel, NMOs grew from 20 μm to 200 μm within 3 days, forming solid or hollow structures (Figure 1A). Enzymatic and mechanical dissociation were utilized for passaging every 7 days, allowing at least 6 months of efficient expansion. Histological analysis using H&E staining (Figure 1A) confirmed that the NMOs closely resembled native tissue. Immunofluorescence staining identified key cell types (17), including epithelial cells (Cytokeratin 7) and ciliated cells (β-tubulin) (Figure 1B).
Growth dynamics of nasal mucosal organoids (NMOs). A, bright field image of NMO at day 1, 3, 5, 7 of passage 0; passage 3, 8, 15, and 25; and the H&E staining (Scale bar = 100 μm); B, immunofluorescence staining of Cytokeratin 7 (CK7), p63, Muc5AC, and β-tubulin. The flow-cytometry results of β-tubulin; C, Muc5AC; D, CK5; E, CD68; F, CD45; G, C5a-R; H, CD3 + CD8 +; I, CD3 + CD4 +; J, of NMO and original tissue.
Flow cytometry revealed a higher percentage of CK7-positive epithelial cells and a lower percentage of CD45-positive lymphocytes (Figure 1C-J), along with an increase in β-tubulin-positive ciliated cells, which are essential for nasal mucosal function (18). Additionally, the NMOs supported immune cells, providing a foundation for further studies on the immune microenvironment.
4.2. Nasal Mucosal Organoids Susceptible to SARS-CoV-2 Pseudovirus and HCoV-OC43 Pseudovirus
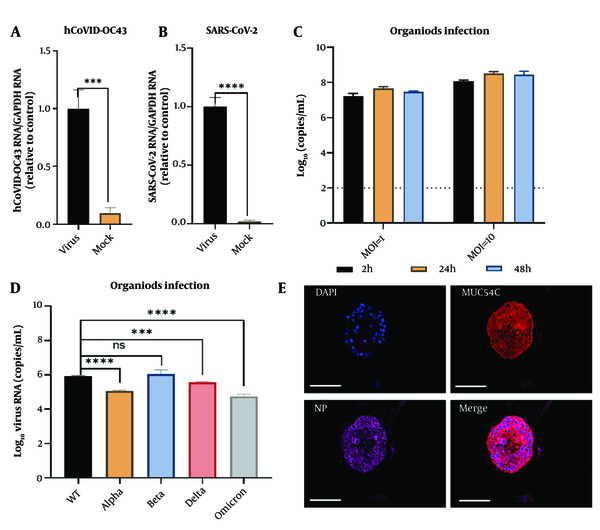
SARS-CoV-2 and HCoV-OC43 pseudoviruses were added to a 96-well plate (30 μL per well) and incubated at 37°C for 2 hours. The supernatant was then replaced with 100 μL of organoid culture medium and incubated for an additional 24 hours. After discarding the medium, the organoids were collected for intracellular viral RNA detection. Viral mRNA expression was significantly higher in the HCoV-OC43 group (P < 0.001) (Figure 2A) and even more elevated in the SARS-CoV-2 group (P < 0.0001) (Figure 2B), confirming the successful establishment of infection models for both viruses in human nasal mucosal epithelial organoids.
Modeling of SARS-CoV-2 infection in nasal mucosal organoids (NMOs). Pseudovirus virus HCoV-OC43 (A); and SARS-CoV2 (B); infection of nasal mucosal organoid; virus content in the supernatant at different times of infection for MOI = 1 and MOI = 10 (C); detection of viral content in the supernatant under different types of virus of infection (D); immunofluorescence detection of goblet cells infected by SARS-CoV-2. nuclei (DAPI), goblet cells (Muc5AC), viral nucleoprotein (NP), and Merge (Scale bar = 100 μm) (E). *** P < 0.001; **** P < 0.0001.
4.3. Assessing Susceptibility of Nasal Mucosal Organoids to SARS-CoV-2 Live Virus
The virus was introduced to NMOs at multiplicities of infection (MOIs) of 1 and 10, followed by a 24-hour incubation. Detection of viral RNA revealed that the MOI = 10 group exhibited higher SARS-CoV-2 copy numbers compared to the MOI = 1 group (Figure 2C), suggesting a correlation between viral exposure and nasal cell susceptibility. Testing various SARS-CoV-2 strains demonstrated similar susceptibility and replication in NMOs (Figure 2D), with viral nucleocapsid primarily expressed in goblet cells, indicating potential sites for viral persistence (Figure 2E).
4.4. Antiviral Drugs Inhibit SARS-CoV-2 Infection in Nasal Mucosal Organoids
The antiviral drug testing capability of the NMO model was evaluated using three antiviral agents: Camostat, remdesivir, and bergamottin. Camostat and remdesivir have established antiviral efficacy, blocking TMPRSS2 and inhibiting RNA-dependent RNA polymerase, respectively (19, 20). Therefore, drug treatments were administered prior to viral introduction to evaluate their preventive efficacy.
Since the adverse effects of bergamottin are unclear, a safety test was conducted first. An ATP assay indicated no significant difference in NMO survival at bergamottin concentrations below 125 μM compared to the DMSO control, consistent with morphological assessments (Figure 3A). H&E staining confirmed the absence of pathological changes or apoptosis at these concentrations (Figure 3B, C).
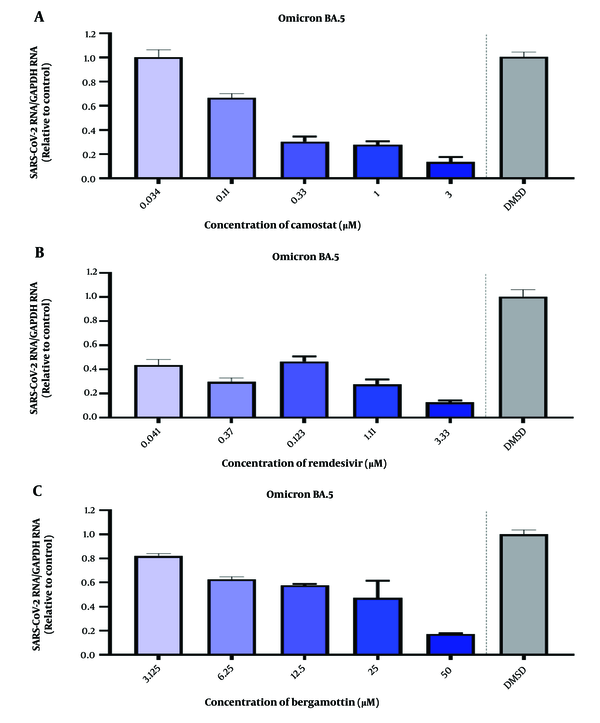
Following the infection of NMOs with SARS-CoV-2 Omicron BA.5 at MOI = 1 for 2 hours, camostat (3 μM) demonstrated dose-dependent inhibition (Figure 4A), while remdesivir reduced viral RNA levels by approximately 40% at 0.041 μM (Figure 4B). Bergamottin showed comparable inhibitory effects, particularly at 50 μM (Figure 4C).
Cell validation evaluation of bergamottin against nasal mucosal organoids (NMOs). A, morphological observation of NMOs treated with 125μM bergamottin scale bar = 1000 μm; B, H&E staining of NMOs under the influence of bergamottin; C, cell viability of NMOs under the influence of different concentrations of bergamottin at 24, 48, and 72 hours. * P < 0.05.
Detection of the inhibitory effect of drugs on SARS-CoV-2 in human nasal mucosal organoids (NMOs). A, camostat at a concentration of 3 μM shows a dose-dependent inhibition of SARS-CoV-2 infection of human NMOs; B, remdesivir at a concentration of 3.33 μM shows a dose-dependent inhibition of SARS-CoV-2 infection of human NMOs; C, bergamottin at a concentration of 50 μM shows a dose-dependent inhibition of SARS-CoV-2 infection of human NMOs.
5. Discussion
Traditional virus research models, such as 2D-cultured immortalized nasal mucosal epithelial cell lines, are limited by their lack of tissue structure and physiological function, as they are derived from monoclonal cell lines. Consequently, they fail to accurately replicate infection processes and pathophysiological changes within the organ. These cell lines also exhibit a uniform genetic background, making them unsuitable for capturing inter-individual variations, thereby restricting their utility in population studies and high-throughput analyses (2, 21-24). In contrast, coronavirus animal models, including rhesus macaques, transgenic mice, and ferrets, present significant physiological disparities compared to humans and are associated with high costs and biosafety risks (3, 25). The nasal cavity, as the primary barrier and initial target organ for respiratory viruses, plays a critical role in viral invasion, replication, and transmission. The nasal mucosa serves as a reservoir for viral replication and a trigger point for immune responses in COVID-19 infection (5). This makes NMOs an ideal model for investigating the mechanisms of COVID-19.
Pseudoviruses, engineered to mimic wild-type viruses, are non-replicating and undergo only a single round of infection, enhancing biosafety while allowing for detection and analysis via reporter genes (26). Both HCoV-OC43 and SARS-CoV-2, classified as β-coronaviruses, can cause a range of diseases in humans and animals (27, 28). Initial experiments using NMOs infected with HCoV-OC43 and SARS-CoV-2 pseudoviruses confirmed the susceptibility of NMOs to coronaviruses. The nasal mucosa, composed of ciliated and goblet cells, exhibits high expression of ACE2 and TMPRSS2, making it the primary site of SARS-CoV-2 infection (29). The virus spreads through goblet cell secretions and ciliary movement (30). Our experiments demonstrated a positive correlation between higher viral exposure doses and increased infection rates and replication. Immunofluorescence analysis revealed that goblet cells are particularly susceptible to SARS-CoV-2, consistent with previous findings (30).
Post-infection, we observed a continuous increase in viral gene copy numbers in NMOs, indicating effective infection and replication. This confirms that NMOs can replicate the complete lifecycle of the virus. The nasal mucosa's large surface area and rich blood supply facilitate efficient drug delivery while minimizing systemic side effects (31-34). Following the COVID-19 outbreak, various nasal-administered antiviral drugs have been developed, yet the lack of robust preclinical models remains a significant bottleneck. The establishment of our human nasal mucosa model for SARS-CoV-2 provides a cost-effective, high-throughput platform for drug evaluation. The primary limitations of this study include a small sample size and the absence of analysis on the variability of viral infection across different populations. Additionally, the antiviral mechanism of bergamottin was not addressed in this study and warrants further investigation.
5.1. Conclusions
Our research demonstrates that camostat, remdesivir, and bergamottin exhibit dose-dependent antiviral properties against SARS-CoV-2 in NMOs, confirming the model’s efficacy as a drug evaluation platform. In conclusion, we have developed a stable in vitro model of SARS-CoV-2 infection using NMOs, which can be utilized for studying infection mechanisms and screening antiviral drugs, offering valuable insights into nasal drug administration for COVID-19 prevention and treatment.