1. Background
Enterococcus faecium is widely distributed in natural environments such as soil and is a normal commensal of the human gut. Initially considered a probiotic (1, 2), it was later recognized as an opportunistic pathogen and removed from the "generally recognized as safe" category (3). Clinically, E. faecium can cause severe infections, including bacteremia, urinary tract infections, and endocarditis. As part of the ESKAPE pathogens (E. faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), a group of bacteria known for their ability to escape the effects of antimicrobial agents, E. faecium is known for its antibiotic resistance (4). Among antibiotic-resistant E. faecium, vancomycin-resistant E. faecium (VREfm) represents a significant public health threat and is included in the high-priority group of the WHO’s Bacterial Priority Pathogens List (2024 update); its prevalence is notably high in certain regions of China (5).
The phylogenetic lineage of E. faecium reflects its health risks and association with hospital infections. Phylogenetic analysis categorizes E. faecium into three clades: A1, A2, and B (E. lactis), with distinct clades showing varying associations with hospital infections. Clade A1 is strongly linked to hospital infections. Research shows that the hospital-associated (HA) clade is generally ampicillin-resistant and exhibits a broader range of antimicrobial resistance compared to the community-associated (CA) clade (6). The HA clade also carries unique sequence variants of microbial surface components involved in matrix adhesion and pilus-encoding genes (7). Colonization-associated proteins (e.g., hyl, ptsD, orf1481) and the genomic plasticity-related sequence IS16 are predominantly found in the HA clade; their distribution can help differentiate clinical E. faecium strains (8).
Inter-clade strain interactions and transitions are essential for the formation of the HA clade. Studies suggest that mobile genetic elements (MGEs) carrying antibiotic resistance genes (ARGs) and virulence factors contribute to the genomic plasticity of E. faecium, facilitating the emergence and spread of new HA clones in clinical settings (9, 10). Soil serves as a natural habitat for E. faecium, with studies indicating that it is the most prevalent Enterococcus species isolated from local soils, exhibiting multi-antibiotic resistance and multi-virulence factors, suggesting that soil may act as a transmission point to humans (11). However, the phylogenetic relationship, resistance, and virulence profiles of hospital soil-derived E. faecium compared to clinical isolates have not been fully explored. This knowledge gap limits understanding of the potential link between hospital soil E. faecium and hospital infections, underestimating its role as a reservoir of HA E. faecium.
2. Objectives
The present study aims to analyze whole-genome sequencing (WGS), resistance phenotypes, and molecular biological characteristics to identify differences and correlations between hospital soil-derived and clinical HA E. faecium. The ultimate goal is to reduce the health risks posed by E. faecium.
3. Methods
3.1. Enterococcus faecium Isolates
This study was conducted at the First Affiliated Hospital of Zhejiang University School of Medicine in Hangzhou, China. Soil samples were collected from the hospital’s roadside greenbelt at 2 - 3 meter intervals from March 6 - 10, 2023. Approximately 0.5 g of each soil sample was collected, enriched in brain heart infusion broth, and plated on Kenner fecal (KF) Streptococcus agar containing sodium azide for incubation at 36°C for 24 hours (12). Suspected colonies with bright red and pink circular appearances were identified using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Identified Enterococcus species were included in the study. Additionally, 31 E. faecium strains, isolated from fecal samples of inpatients between September 13, 2010, and September 12, 2019, at the same hospital, were also included.
3.2. Antibiotic Susceptibility Test
All isolates underwent antibiotic susceptibility testing (AST) using broth and agar dilution methods based on CLSI guidelines, 33rd edition, 2023 (13). Broth dilution tested vancomycin, teicoplanin, and tigecycline, while agar dilution tested minocycline, tetracycline, ciprofloxacin, levofloxacin, ampicillin, penicillin, erythromycin, rifampicin, linezolid, and chloramphenicol. Results were interpreted according to CLSI parameters, with E. faecalis ATCC 29212 as the quality control strain.
3.3. Whole-Genome Sequencing, Assembly, and Data Analysis
Genomic DNA was extracted using the FastDNA® Spin Kit for Soil (MP Biomedicals, Illkirch, France). The WGS was performed on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA). The quality of raw sequencing reads was assessed using FastQC v.0.11.5, and adapters were trimmed with Trimmomatic v.0.40 (https://www.bioinformatics.babraham.ac) (14). Trimmed reads were assembled de novo with SPAdes v.3.6 (15). Genome annotation was performed using the RAST server (rast.nmpdr.org) and Prokka (16, 17). The Next generation sequencing (NGS) data are available in the NCBI Genome repository under accession number PRJNA1141267, with additional dataset accession numbers listed in Appendix 1 in Supplementary File (available online).
3.4. Phylogenetic Analysis
Phylogenetic relationships between hospital soil-derived and clinical E. faecium strains were analyzed to validate clade assignments. Fifty-six E. faecium strains, along with three reference strains representing the HA (Efm TX16, GCF_000174395.2), CA (Efm C59, GCF_009707505.1), and Els (Efm TX1330, GCF_003583905.1) clades, were included. Phylogenetic analysis was performed using kSNP4 (18) based on core genome single nucleotide polymorphisms (cgSNP) with default parameters (K = 15) to construct a neighbor-joining tree. The tree was visualized using iTOL V6 (19), and annotations from other molecular analyses were integrated. Panaroo v1.1.31 (20) extracted the core genome from all strains, aligned using SNP-sites v2.5.1 (21). A minimum spanning tree (MST) was reconstructed with PHYLOViZ v2.01 and the geoBURST Full MST algorithm (22).
3.5. Classification of Sequences
The 56 E. faecium strains were grouped based on informative SNPs using RhierBAPS software (23), with a maximum depth of 2 and a maximum of 20 populations. Multi-locus sequence typing (MLST) was performed using the unpublished mlst software by Seemann (https://github.com/tseemann/mlst). For strains without sequence type (ST) assignments, new STs were assigned by submitting the sequences to the pubMLST database (24).
3.6. Identification of Antibiotic Resistance and Virulence Genes, Plasmids, and Transposons
The ARGs were identified using CGE ResFinder 4.5.0 (25) with a similarity threshold of 90% and a minimum length of 60%. Virulence factor genes were detected using CGE VirulenceFinder 2.0 (26) under the same thresholds for Enterococcus and E. faecium and E. lactis. Plasmid replicons were identified using CGE PlasmidFinder 2.1 (27) with the database set to gram positive. Antibiotic resistance-related MGEs were analyzed using VRprofile2 (28) at the single genome contig level.
3.7. Population Structure Analysis
The isolates underwent admixture analysis using ADMIXTURE version 1.3.0 (29). We tested the number of ancestral populations (K) from one to six and eventually set it at three. The alignment data were converted to BED format using PLINK 1.90 beta (30).
3.8. Statistical Analysis
Statistical analysis was performed using R version 4.4.0; a P-value ≤ 0.05 was considered statistically significant. Data from different groups were compared using the Kruskal-Wallis test.
4. Results
4.1. Epidemiologic Analysis of Enterococcus faecium
Among 320 isolation sites, we identified 25 isolates (Figure 1) of E. faecium using MALDI-TOF-MS. The predominant STs were ST555 (five isolates), ST32 (three isolates), ST2608 (three isolates), and ST2609 (three isolates). We identified four new STs: ST2607 (one isolate), ST2608 (three isolates), ST2609 (three isolates), and ST2610 (one isolate). Furthermore, we isolated 31 E. faecium strains from patient fecal samples in the same hospital. The MLST analysis showed the predominant STs were ST78 (12 isolates), ST341 (six isolates), ST17 (three isolates), and ST555 (three isolates).
A sketch map of sampling soil sites within the hospital environment. The small black dots represent all sampled sites, while larger dots represent those sites where Enterococcus faecium strains were isolated. Genetically linked isolates [core genome single nucleotide polymorphisms (cgSNP) alleles ≤ 25] are depicted by dots with the same color.
4.2. Phylogenetic Analysis and Genomic Characteristics of Enterococcus faecium
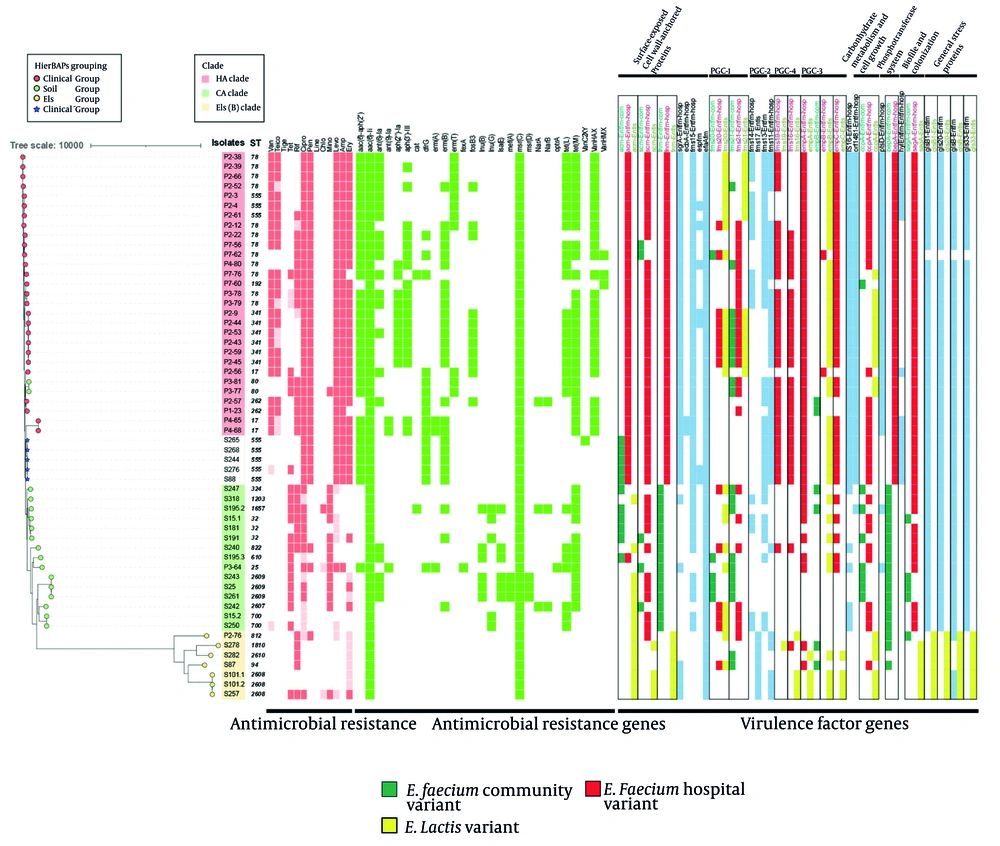
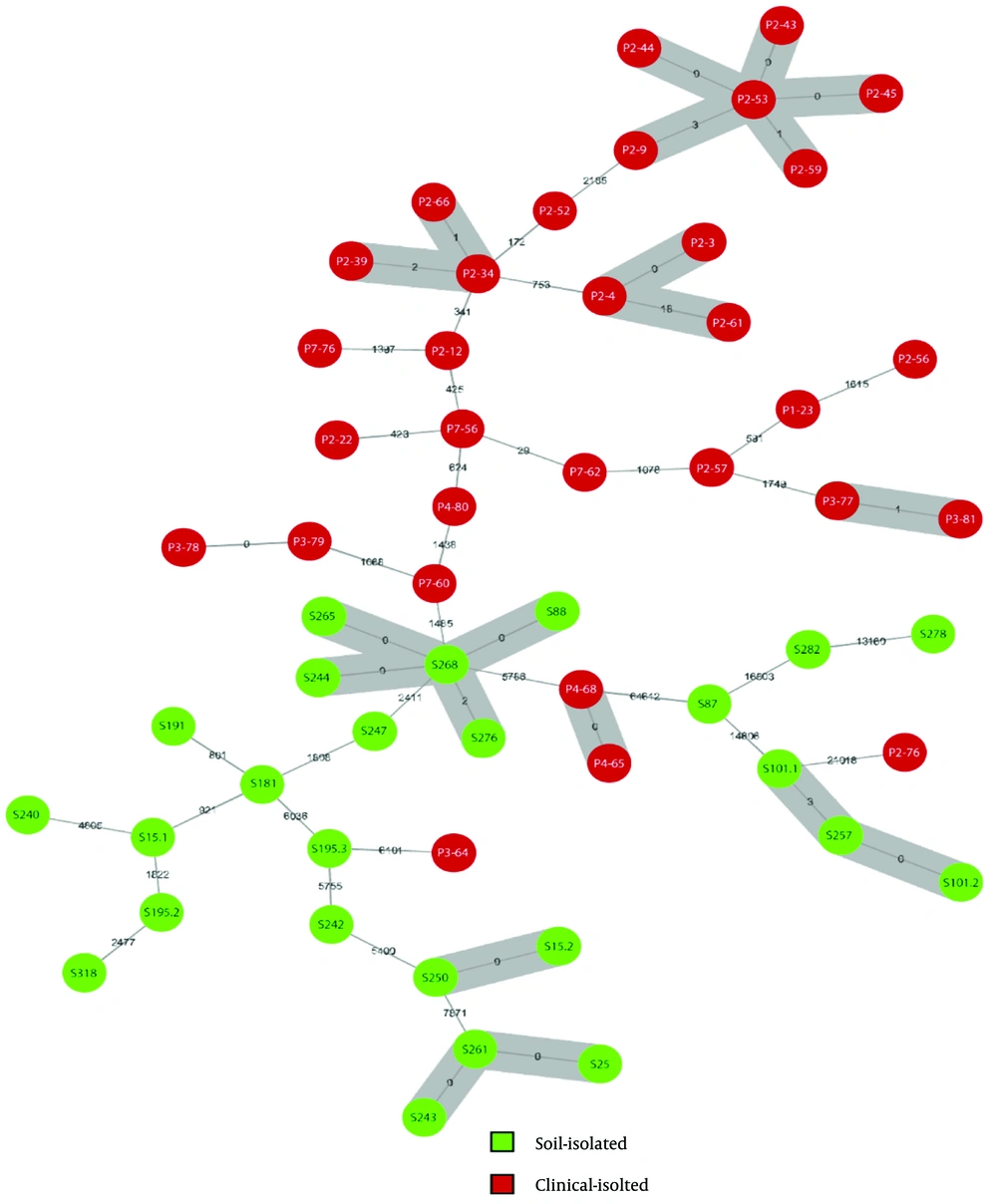
Phylogenetic relationships of hospital soil-derived and clinical E. faecium isolates were analyzed to verify clade distribution. A neighbor-joining tree based on core genome SNPs from 56 isolates and three reference strains was constructed (Figure 2), dividing isolates into three clades: The HA clade reference strain (Efm TX16, GCF_000174395.2) grouped exclusively with clinical isolates, the CA clade reference strain (Efm C59, GCF_009707505.1) grouped mainly with soil isolates except for one clinical isolate, and the Els clade reference strain (Efm TX1330, GCF_003583905.1) grouped within the Els clade. Using RhierBAPS, SNPs were further grouped into four groups (Figure 2). Group 1 (clinical group) included 27 clinical isolates; group 2 (Els group) comprised seven E. lactis isolates; and group 3 (soil group) contained 17 isolates, mostly soil-derived (14 soil, three clinical). Five soil strains from group 4, forming a distinct cluster within the clinical clade, were designated the “clinicalʹ group”. These strains may represent a transitional component between HA and CA E. faecium, potentially persisting in hospital soil while maintaining traits linked to clinical infections. To assess relatedness and transmission between clinical and hospital soil environments, we constructed a MST based on core genome SNPs (Figure 3). Using a threshold of ≤ 25 cgSNP alleles to define relatedness (31), we identified six genetically related clusters among 18 clinical isolates and four clusters among 13 soil isolates. No direct transmission between clinical and soil environments was detected; however, the clustering of the clinicalʹ group suggested notable dissemination within the hospital soil environment, impacting both the eastern and western areas.
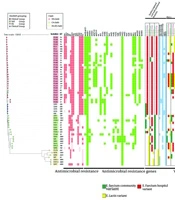
A heatmap was generated showing antibiotic susceptibility, antibiotic resistance gene (ARG) distribution, and virulence factor gene variants. Isolate labels indicate their source: P for patients, S for soil. Clade assignments are represented by colored strips, while HierBAPS-based groupings are shown with different colored symbols. Dark red indicates resistance, and lighter red indicates intermediate resistance. The antibiotics tested include: Vancomycin (Van), teicoplanin (Teico), tigecycline (Tige), tetracycline (Tet), rifampicin (Rif), ciprofloxacin (Cipro), penicillin (Pen), linezolid (Line), chloramphenicol (Chlo), minocycline (Mino), levofloxacin (Levo), ampicillin (Amp), and erythromycin (Ery). Variants of virulence factor genes are boxed, with color codes: Green for the Enterococcus faecium community variant, red for the hospital variant, and yellow for the E. lactis variant.
The minimum spanning tree (MST) based on core genome SNPs of 56 isolates. Each circle represents an isolate, with green circles representing soil isolates and red circles representing clinical isolates. The numbers between isolates show their core genome single nucleotide polymorphisms (cgSNP) distance. Isolates within the threshold of 25 alleles are considered genetically linked; gray zones were drawn to bridge these isolates.
4.3. Antibiotic Susceptibility Results
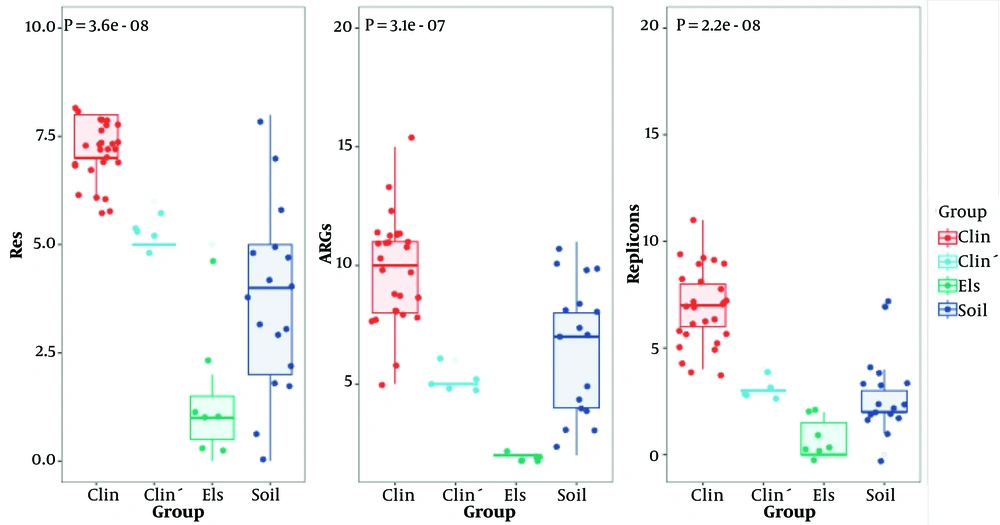
To assess the transitional role of the clinicalʹ group, antibiotic resistance phenotypes of isolates from different sources were compared. The AST revealed that the clinical group exhibited significantly more resistance types than the soil and Els groups (7.11 ± 0.70, 3.76 ± 2.11, 1.43 ± 1.72, and P < 0.001) (Figure 4). The clinical group was predominantly resistant to vancomycin, teicoplanin, erythromycin, ciprofloxacin, penicillin, levofloxacin, and ampicillin. Interestingly, the clinical group commonly displayed a resistance profile including ciprofloxacin, penicillin, levofloxacin, ampicillin, and erythromycin, while the soil group isolates showed no such profile. However, the clinicalʹ group carried the same antibiotic resistance profile, only lacking resistance to vancomycin and teicoplanin.
To further distinguish the clinicalʹ, clinical, and soil groups, molecular features were analyzed using WGS data. Among 27 types of ARGs detected, the clinical group exhibited significantly more ARGs than the soil and Els groups (9.85 ± 2.11, 6.53 ± 2.85, 2.00 ± 0.00, P < 0.001) (Figure 3). The clinicalʹ group was comparable with the soil group in overall ARG number but had significantly fewer ARGs than the clinical group. However, the clinicalʹ group carried ARGs such as aac(6')-aph(2''), dfrG, and erm(B), which were more commonly found in the clinical group. These ARGs likely contribute to the resistance profile shared with the clinical group. Although the clinicalʹ group resembled the soil group in overall ARG number, its possession of certain ARGs typically found in the clinical group was notable.
To determine if ARGs were associated with MGEs, the VRprofile2 database was used. Seven MGEs associated with single resistance genes and one MGE linked to multiple resistance genes were identified. The aac(6')-aph(2'') + IS1216V combination was the only MGE-associated ARG found in both clinical and soil isolates, specifically represented by soil strains in the clinicalʹ group.
Virulence profiles of HA and CA E. faecium clades were compared using VirulenceFinder (Figure 2). Among 30 virulence factor genes, acm, fnm, fms-19, fms-16, and bepA showed strong source-specificity. Most clinical group strains exhibited HA variants, and soil group strains displayed CA variants, while the Els group exclusively showed Els variants. Similar to the clinical group, the clinicalʹ group exhibited primarily HA variants of these genes, though it carried CA variants for the acm gene. No other CA or Els variants were detected. Genes such as sgrA, fms11, IS16, orf1481, ptsD, and hyl were predominantly found in the clinical group, with limited presence in the Els group and almost none in the soil group. The clinicalʹ group carried all these genes except for ptsD. Notably, the ptsD gene was present in nearly all clinical strains (30/31) but only one soil strain (1/25), demonstrating its specificity for clinical isolates. In the clinicalʹ group, surface and pilus protein sequences were primarily HA variants. This group lacked the PGC-1 (pilus gene cluster) gene, had an incomplete PGC-2 gene, and exhibited most HA putative virulence markers (PVMs). The clinicalʹ group’s virulence factor distribution closely matched that of the HA group, indicating nearly complete pathogenicity.
Plasmid carriage significantly contributes to the genomic plasticity of E. faecium. We used PlasmidFinder to detect the number of replicons in the 56 strains. A total of 20 replicons were detected, with the clinical group exhibiting significantly more replicons than the soil and Els groups (6.85 ± 1.79, 2.82 ± 1.85, 0.71 ± 0.95, P < 0.001) (Figure 3). The Rep11a replicon was unique to the clinical and clinicalʹ groups.
4.4. Population Structure Analysis
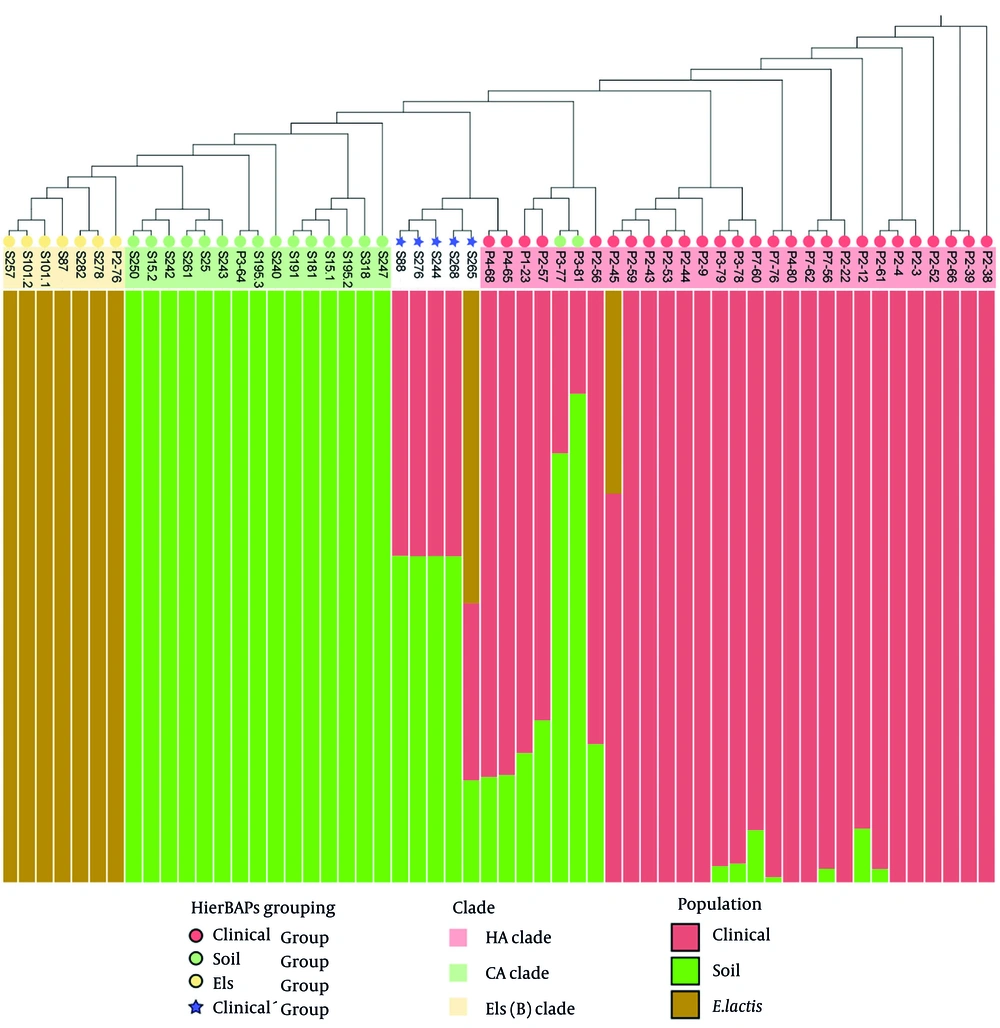
To elucidate the evolutionary contributions of clades of E. faecium, we used admixture analysis to define ancestral populations based on genetic characteristics and allocate individuals within the population. Considering the previously discussed clade distributions and sequence groupings, we hypothesized that all strains originated from three ancestral populations. Our findings (Figure 5) indicate that the ancestral components of the CA and Els clades are respectively homogeneous, without contributions from other populations. Naturally, the two populations were assigned as soil and E. lactis ancestral components; the predominant ancestral component of the HA clade was assigned as the clinical ancestral component. Several strains within the HA clade possessed soil ancestral components; one strain had Els ancestral components. Two clinical E. faecium strains previously categorized into the soil group by hierBAPS displayed greater soil ancestral contributions than the clinical one, validating the accuracy of our grouping and suggesting interactions between HA and CA E. faecium clades in hospital soil and clinical settings. Notably, four strains in the clinicalʹ group contained comparable clinical and soil ancestral components; one strain had clinical, soil, and E. lactis ancestral components, with nearly equivalent contributions from clinical and soil ancestries. This further supports the hypothesis that the clinicalʹ group represents a continuum bridging HA and CA E. faecium clades.
5. Discussion
In this study, clinical isolates of E. faecium comprised the HA clade, while isolates from hospital soil environments constituted the CA and Els clades, consistent with common multi-source E. faecium phylogenetic distributions. A significant trend was observed with ARGs and replicons in each group decreasing from the clinical group to the soil and Els groups, suggesting that environmental E. faecium isolates have lower resistance rates to antibiotics and fewer overall virulence genes. This supports the association between the role of ARGs and replicons and the clinical adaptation of E. faecium.
Considering this association, we explored and identified HA clade-related clinicalʹ group E. faecium in hospital soil environments based on their phenotypical and molecular biological comparative analysis. Aligning with the HA clade, the clinicalʹ group is distinguished from the CA clade by its shared resistance spectrum, almost identical virulence profile with the HA clade, and population structure analysis showed that the ancestral components of HA and CA clades contributed equally to the clinicalʹ group’s genetic composition. Previous research has shown that the predecessor of the current vancomycin-resistant E. faecium (VREfm) was the HA clade ampicillin-resistant E. faecium (AREfm), which evolved into VREfm through horizontal gene transfer of the van gene (32). The clinicalʹ group E. faecium identified in this study also exhibited ampicillin resistance, a trait absent in the CA clade but predominant in the HA clade. Therefore, we believe that the clinicalʹ group E. faecium parallels historical AREfm in characteristics, with the potential of becoming VREfm by acquiring van genes during infection or colonization in clinical settings.
The resistance phenotype results suggest that ampicillin resistance is not the only differentiation. This study identified a set of antibiotic resistances present only in the clinical and clinicalʹ groups. We assume that such resistance is essential for colonization in patients amidst exposure to various antibiotics in clinical settings, conferring an advantage and resilience against elimination during subsequent treatments. Soil strains lack this uniform resistance distribution, highlighting its significance in the evolutionary adaptation from the CA to the HA E. faecium clades. Notably, the distribution of soil-derived strain clusters in the MST indicates cross-regional transmission in hospital soil, with three clusters, including the clinicalʹ group, spanning from the east to the west of the hospital. This proves that soil E. faecium can traverse long distances with carrier assistance and re-establish in new soil, posing potential health risks due to their similarity to HA E. faecium.
Soil is recognized as a reservoir for ARG-carrying pathogens (33). Likewise, soil-dwelling E. faecium can accept resistance genes like tet(M), erm(B), erm(Q), and mef(A) from Clostridium perfringens, demonstrating that resistance genes in soil can transfer to Enterococcus species (34). Their ability to exchange mobile elements with other soil bacteria may enable E. faecium to acquire resistance phenotypes or other clinically rare traits. In this study, the mobile element IS1216V combined with the resistance gene aac(6')-aph(2'') in the clinical and clinicalʹ groups may be involved in such processes. Compared to the clinicalʹ and soil groups, the greater diversity of replicons in the clinical group may contribute to or result from its ultimate adaptation to clinical environments. The clinicalʹ group potentially evolves into HA E. faecium by acquiring HA mobile elements through infection or colonization in clinical settings or soil environments.
Studies indicate that genetic exchange across E. faecium clades predominantly occurs from clade B (Els clade) or clade A2 (soil clade) to clade A1 (clinical clade) (10). This explains why the HA clade contains ancestral components from the other two clades, while these two clades lack HA clade components. We acknowledge that the 13-year time interval between clinical and hospital soil isolates may introduce temporal confounders when comparing resistance profiles. However, longitudinal surveillance studies have demonstrated that the resistome of E. faecium, particularly hospital-adapted strains, remains relatively stable over time. For example, surveillance data from Italy showed minimal fluctuations in resistance rates to key antibiotics such as ampicillin, linezolid, teicoplanin, tigecycline, and vancomycin across multiple years (35). Similarly, national data from China’s BRICS program (2016 - 2022) (36-40) confirmed consistent resistance rates for these agents among clinical E. faecium isolates. Within our own dataset, we found no significant differences in resistance phenotypes or resistance gene profiles among patient-derived isolates collected between 2010 and 2019. This internal consistency suggests that, at least within the studied hospital setting, the confounding effect of temporal separation is likely minimal.
We also acknowledge that bacterial resistance mechanisms are dynamic, and that CLSI guidelines for antimicrobial susceptibility testing are updated annually to reflect these changes. However, the antibiotics selected in this study — including ampicillin, penicillin, ciprofloxacin, levofloxacin, erythromycin, vancomycin, and teicoplanin — have consistently remained among the primary agents for E. faecium susceptibility testing over the past decade. Consequently, despite the time interval between patient and soil isolate collections, the major resistance phenotypes compared in this study reflect stable and clinically relevant antibiotic targets. Moreover, our analysis focused on well-established resistance traits rather than newly emerging or rare mechanisms, minimizing the confounding impact of evolving resistance patterns on the validity of our comparisons.
5.1. Conclusions
In conclusion, we isolated five transitional strains from the hospital soil environment. Comparative analyses of resistance phenotypes, ARG distribution, and virulence profiles demonstrate that these strains are transitional between HA and CA E. faecium clades, suggesting their potential for clinical infection. Population structure analysis further showed that the HA and CA clades contributed equally to the ancestral components of the clinicalʹ group, making it a continuum bridging the two clades. The emergence of the clinicalʹ group supports the adaptive transformation process between CA and HA clades and demonstrates that hospital soil environments are not as safe as previously thought. Clinically hazardous E. faecium can adapt to and exploit the hospital soil as a reservoir, facilitating its spread over long distances within hospital environments.
![A sketch map of sampling soil sites within the hospital environment. The small black dots represent all sampled sites, while larger dots represent those sites where <i>Enterococcus faecium</i> strains were isolated. Genetically linked isolates [core genome single nucleotide polymorphisms (cgSNP) alleles ≤ 25] are depicted by dots with the same color. A sketch map of sampling soil sites within the hospital environment. The small black dots represent all sampled sites, while larger dots represent those sites where <i>Enterococcus faecium</i> strains were isolated. Genetically linked isolates [core genome single nucleotide polymorphisms (cgSNP) alleles ≤ 25] are depicted by dots with the same color.](https://services.brieflands.com/cdn/serve/3170b/7088995bc800abb3cb1190c72716bcf8dbffd915/jjm-159860-i001-F1-preview.webp)