1. Background
In late December 2019, a new virus from the Coronavirus family was detected in Wuhan, China, which quickly spread globally (1, 2). On March 11, 2020, the World Health Organization (WHO) declared the outbreak of COVID-19 a pandemic (3). The virus was named "SARS-CoV-2" by the International Committee on Taxonomy of Viruses (ICTV) (4). SARS-CoV-2 shares 80% phylogenetic similarity with SARS-CoV and 50% with MERS-CoV, all belonging to the beta coronaviruses genus (5, 6). Understanding the molecular mechanisms underlying the immune response to SARS-CoV-2 is crucial for developing effective treatments and diagnostic tools. Fourteen open reading frames (ORFs) code for both structural and non-structural proteins, such as nucleocapsid (N), membrane (M), envelope (E), and spike (S) proteins (7). By attaching to the ACE2 receptor on host cells, the S protein, which is made up of the S1 and S2 subunits, promotes viral entry (8-10).
Small non-coding RNAs known as microRNAs (miRNAs) are involved in the regulation of gene expression post-transcriptionally, playing crucial roles in cellular functions, disease mechanisms, immune response, and viral infections. The miRNAs have been identified as potential diagnostic tools and therapeutic targets in various diseases, including viral infections (11). The miRNAs are isolated from blood (serum and plasma), saliva, urine, feces, and other body fluids (12). Several new methods have been developed with a special focus on sensitivity and specificity for the recognition of miRNAs in human body fluids (13). Current methods used for detection include quantitative PCR (qPCR), in situ hybridization, microarrays, and RNA sequencing (14).
Recent studies have highlighted the importance of miRNAs in the pathogenesis of viral infections, including their roles in modulating host immune responses and viral replication. However, the specific roles of miRNAs in the context of COVID-19 remain poorly understood. Among the miRNAs, miR200b-3p, miR34b-5p, miR203a-3p, and Let7g-5p have been implicated in inflammatory processes and immune regulation, suggesting their potential involvement in COVID-19 pathophysiology (15, 16).
MiR200b-3p is a miRNA that can regulate gene expression and influence the expression of ACE2 in COVID-19 patients (17). It can also modulate the level of interleukin-6 (IL-6) and other cytokines (18). MiR203a-3p can influence the expression of genes involved in inflammation, potentially affecting the severity of COVID-19 disease (19). MiR34b-5p can interact with key proteins and immune response pathways (20), and Let7g-5p is known to inhibit M protein expression of SARS-CoV-2 and suppress the immune response (21).
The IL-6 is a pro-inflammatory cytokine identified as a key marker of severe COVID-19. Elevated levels of IL-6 are associated with increased disease severity and poor clinical outcomes in COVID-19 patients. Understanding the relationship between miRNA expression and IL-6 levels could provide insights into the molecular mechanisms driving inflammation and disease progression in COVID-19 (22, 23). Despite the growing body of literature on miRNAs and IL-6 in viral infections, there is a lack of comprehensive studies examining their roles and interactions in COVID-19 patients. This study aims to investigate the expression levels of miR200b-3p, miR34b-5p, miR203a-3p, and Let7g-5p in COVID-19 patients and their correlation with IL-6 expression levels.
2. Objectives
We hypothesize that these miRNAs are up-regulated in COVID-19 patients and correlated with IL-6 levels, contributing to the inflammatory response observed in severe cases.
3. Methods
3.1. Characteristics of the Subjects
Sixty hospitalized COVID-19 patients (26 females, 34 males) confirmed by specialists and real-time PCR, along with 30 healthy individuals, were included in this study. Patients were admitted to the infectious disease ward of Razi Hospital (Ahwaz, Iran) between May and July 2021. The O2 saturation of all patients was ≤ 93%, categorizing them as severe, and they received remdesivir (first day 200 mg/IV, second to fifth days 100 mg/IV) based on the national therapeutic protocol. Thirty healthy individuals with no signs of disease and a negative real-time PCR test were included as the control group. Sample collection was carried out on the first day of admission, and written informed consent was obtained from all participants. Demographic information was collected via a questionnaire.
Exclusion criteria for patients included a history of asthma, allergies, autoimmune, or inflammatory diseases. Healthy controls were screened to exclude any history of COVID-19 symptoms or recent exposure to infection, confirmed history of SARS-CoV-2 infection by PCR test, other respiratory infections, and chronic illnesses that could impact miRNA expression. Additionally, they were not taking immunosuppressants or anti-inflammatory medications, and none were pregnant.
3.2. Patient Management
Blood samples and nasopharyngeal swabs were collected on the first day of the visit. Dacron-flocked swabs (Good Care, China) were used for collecting nasopharyngeal samples following standard protocols. Samples were immediately placed in a viral transport medium and transferred to the virology laboratory under cold chain conditions. For separating PBMCs, Ficoll-Paque density gradient centrifugation was carried out. Plasmas and PBMCs were stored at -70°C for further analysis.
3.3. Gene Expression
Total RNA was extracted from plasma samples using the SinaPure™ Viral Kit (SinaClon, Iran) according to the manufacturer's instructions. The concentration and purity of RNA were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). cDNA was synthesized from 1 µg of total RNA using the MystiCq miRNA cDNA synthesis kit (Sigma-Aldrich, USA) following the manufacturer's protocol. Real-time PCR was used to evaluate miR200b-3p, miR34b-5p, miR203a-3p, and Let7g-5p expression, with U6 as an internal control. Primers were obtained from Bonyakhteh Company (Iran).
3.4. Real-time PCR for MicroRNA Expression
The expression levels of miR200b-3p, miR34b-5p, miR203a-3p, and Let7g-5p were quantified using the SYBR Green method (Ampliqon, Denmark). U6 small nuclear RNA was used as an internal control. Real-time PCR was performed on an ABI StepOne real-time PCR System (Applied Biosystems, USA) with the following cycling conditions: 95°C for 15 minutes, followed by 40 cycles of 95°C for 20 seconds, 55°C for 20 seconds, and 72°C for 2 seconds. The relative expression levels were calculated using the 2-ΔΔCt method.
3.5. Interleukin-6 Gene Expression and Plasma Concentration
PBMC samples were extracted using the Super RNA Extraction Kit (Ana Cell, Iran). The cDNA synthesis was done using a Sinacolon cDNA synthesis kit (Iran), and IL-6 expression was quantified by real-time PCR using SYBR Green master mix (Ampliqon, Denmark) with GAPDH as an internal control. The following primers were used:
A. IL-6 forward: 5'-ACAATACCCCCAGGAGAAG-3'.
B. IL-6 reverse: 5'-GTCGAGGATGTACCGAATTT-3'.
C. GAPDH forward: 5'-TGTGGGCATCAATGGATTTGG-3'.
D. GAPDH reverse: 5'-ACACCATGTATTCCGGGTCAAT-3'.
The thermal cycling profile was 95°C for 15 minutes, followed by 40 cycles of 95°C for 20 seconds, 55°C for 20 seconds, and 72°C for 20 seconds. IL-6 plasma concentrations were analyzed using kits from Karmania Pars Gene Company, Iran, according to the manufacturer's protocol.
3.6. Data Analysis
Statistical analyses were performed using SPSS software version 22.0 (IBM Corp., USA). The Mann-Whitney U test was used to compare the expression levels of IL-6 and miRNA between COVID-19 patients and controls. Correlations between miRNA and IL-6 expression levels were evaluated using Spearman's correlation coefficient. Statistical significance was defined as a P-value of less than 0.05.
4. Results
The study analyzed the expression levels of four miRNAs and IL-6 in COVID-19 patients compared to controls. The mean ages were 52.69 ± 14.1 years for the patients and 44.75 ± 13.09 years for the controls. Demographic data and comorbidities of COVID-19 patients are mentioned in Table 1.
| Variables | Patients (N = 60) | Controls (N = 30) |
|---|---|---|
| Demographic | ||
| Age (y) | 52.69 ± 14.1 | 44.75 ± 13.09 |
| Sex (male) | 34 (56.66) | 15 (50) |
| Comorbidities | ||
| Hypertention | 8.3 | - |
| Diabetes mellitus | 6.6 | - |
| Hyperlipidemia | 3.3 | - |
| Chronic pulmonary disease | - | - |
| Chronic ischemic heart disease | - | - |
| Arthritis rheumatoid | - | - |
| Hypotyroidism | - | - |
| Mortality rate | - | - |
a Values are expressed as mean ± SD or No. (%).
4.1. MicroRNA Expression Levels
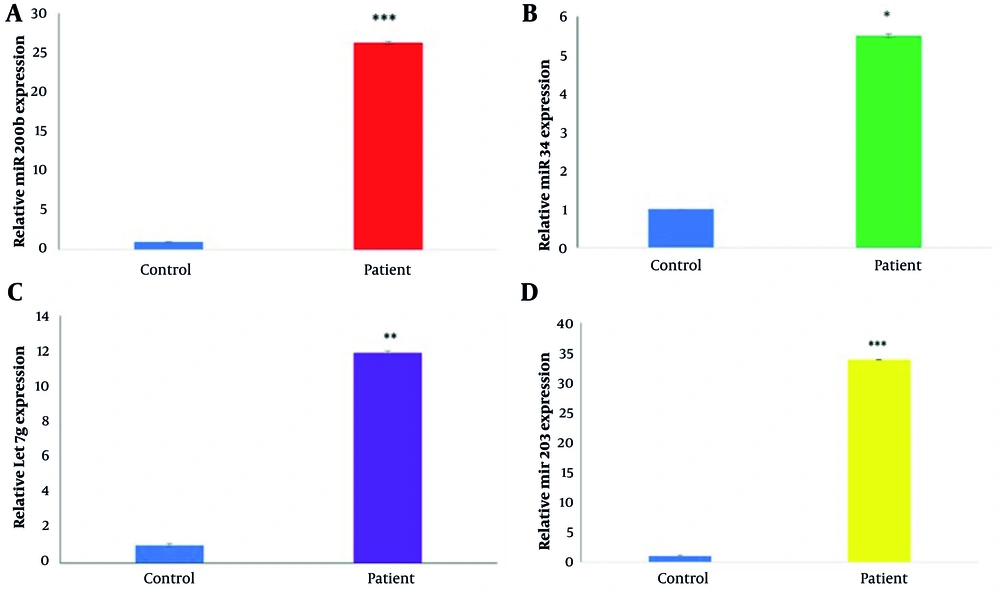
The expression levels of miR200b-3p (26.32-fold change), miR34b-5p (5.51-fold change), miR203a-3p (33.84-fold change), and Let7g-5p (11.93-fold change) were found to be significantly higher in COVID-19 patients than in the control group (Figure 1).
A, comparison of miR200b-3p expression levels in patients vs. control group (P < 0.001); B, comparison of miR34b-5p expression levels in patients vs. control group (P < 0.02); C, comparison of Let7g-5p expression levels in patients vs. control group (P < 0.003); D, comparison of miR203a-3p expression levels in patients vs. control group (P < 0.001). *, P < 0.02; **, P < 0.003; ***, P = 0.00.
4.2. Interleukin-6 Expression Levels
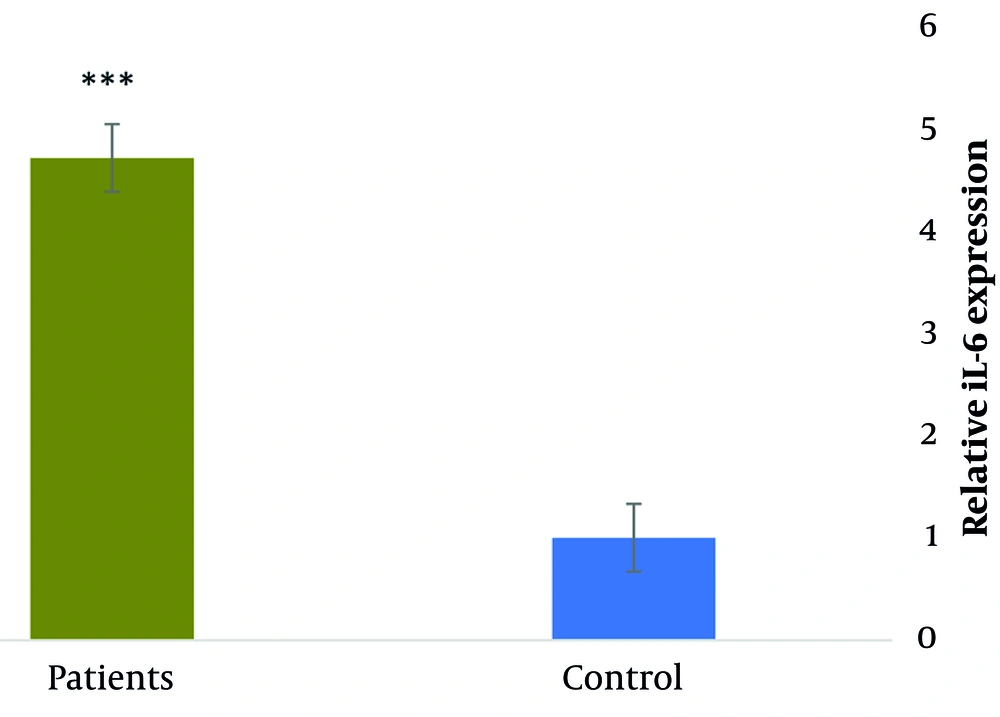
Comparing COVID-19 patients to the control group, there was a significant increase in IL-6 expression (4.7-fold change, P < 0.001). The serum level of IL-6 in COVID-19 patients was 17.34 ± 0.32 pg/mL, which was significantly higher compared to the healthy control group at 1.5 ± 0.23 pg/mL (P < 0.001) (Figure 2).
4.3. Correlation of MicroRNAS with Interleukin-6 Expression
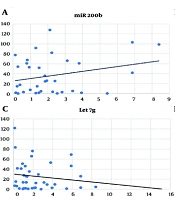
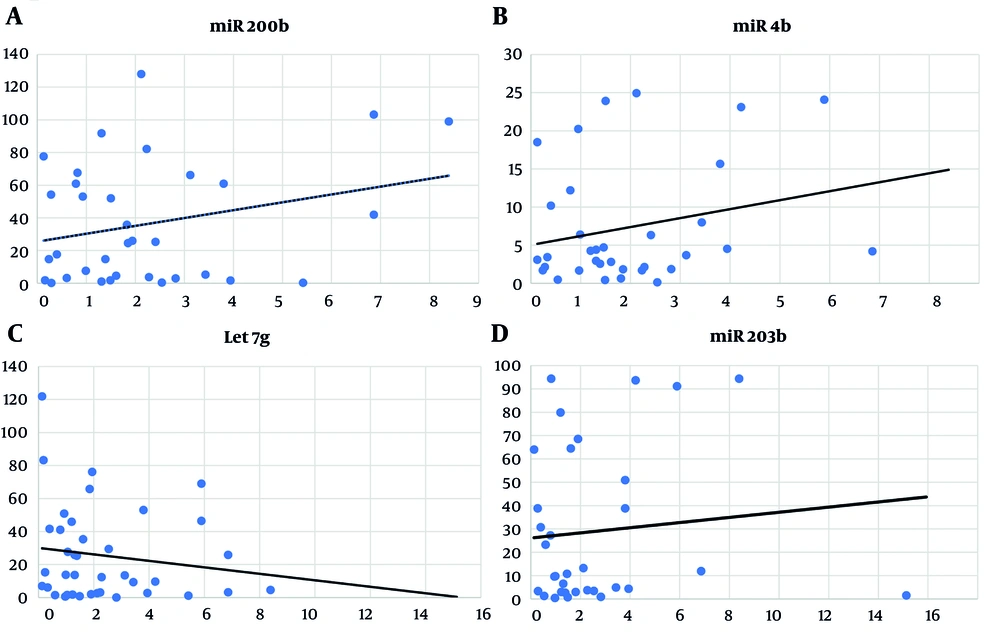
Positive correlations, though not significant, were found between the expression of miR200b-3p, miR34b-5p, miR203a-3p, and IL-6. Conversely, a negative, non-significant correlation was observed with Let7g-5p (Figure 3).
A, miR-200b-3p and interleukin-6 (IL-6) expression correlation analysis in COVID-19 patients (Spearman's correlation = 0.08, P < 0.63); B, correlation analysis between miR-34b-5p and IL-6 expression in COVID-19 patients (Spearman's correlation = 0.139, P < 0.43); C, correlation analysis of Let7g-5p and IL-6 expression in COVID-19 patients (Spearman's correlation = -0.135, P < 0.41); D, correlation analysis of miR-203a-3p and IL-6 expression in COVID-19 patients (Spearman's correlation = 0.06, P < 0.71).
5. Discussion
In comparison to the control group, our study demonstrated significantly elevated levels of miR200b-3p, miR34b-5p, miR203a-3p, Let7g-5p, and IL-6 in the blood of COVID-19 patients. These findings align with those observed in viral infections and underscore their potential roles in the pathophysiology of COVID-19. Notably, COVID-19 patients have higher expression levels of miR200b-3p (24). Previous studies have shown that miR200b-3p is involved in the regulation of inflammatory pathways, suggesting its role in the heightened inflammatory response seen in COVID-19 (25). However, some studies demonstrated that this miRNA's ability to influence cytokine production through mechanisms like the modulation of NF-κB signaling pathways could explain its association with disease manifestations (26).
Similarly, the elevated levels of miR-34b-5p and miR-203a-3p observed in our study align with their known roles in immune response regulation. Notably, miR-34b-5p is predominantly expressed in lung tissue. Its levels have been shown to increase during acute lung injury (ALI), coinciding with a significant reduction in progranulin (PGRN) expression. This upregulation of miR-34b-5p may contribute to the pathogenesis of ALI by downregulating PGRN. In the context of COVID-19, miR-34b-5p might exacerbate inflammation and tissue damage by contributing to a dysregulated immune response (27).
MiR203a-3p overexpression, on the other hand, has been shown to strongly promote H1N1 infection by targeting the host’s IFN signaling pathways and also targeting the viral genome (28). Another in silico study predicted that miR203a-3p targets differentially expressed genes (DEGs) and is involved in SARS-CoV-2 processes (29). Srivastava et al. predicted miR-203a-3p upregulation in severe COVID-19, which is consistent with our study (30). The upregulation of Let7g-5p in COVID-19 patients adds another layer of complexity (21). The altered expression of Let7g-5p could influence the immune response to SARS-CoV-2, contributing to the observed cytokine storm and severe clinical outcomes (31).
Our study also highlights a non-significant positive correlation between the expression levels of these miRNAs and IL-6. The IL-6 is a well-established marker of severe COVID-19, associated with increased disease severity and poor clinical outcomes (32). The observed correlation suggests that these miRNAs might regulate IL-6 production, thereby influencing the inflammatory milieu in COVID-19 patients. This relationship provides valuable insights into the molecular mechanisms driving the inflammatory response in COVID-19 and suggests that these miRNAs may serve as biomarkers for disease severity (33).
Let-7g binds to the target mRNA's 3' untranslated region (UTR) to either block translation or cause mRNA degradation, performing its regulatory role (34). Our study aligns with previous research demonstrating that let-7g targets IL-6 mRNA, reducing its stability and translation, thereby downregulating IL-6 expression (34). The ability of let-7g to modulate IL-6 expression is particularly relevant in the context of COVID-19, where dysregulated cytokine production is a hallmark of severe disease. By negatively regulating IL-6, let-7g may help to prevent the excessive inflammatory response associated with severe COVID-19 (35).
In comparison with existing literature, our findings are consistent with studies that have identified miRNAs as critical regulators of the immune response in viral infections (36). For instance, recent research has shown that miRNAs can directly or indirectly modulate cytokine production, supporting our observations of the positive correlations between miRNA expression and IL-6 levels (37, 38). These findings contribute to the growing body of evidence that miRNAs play pivotal roles in the pathogenesis of COVID-19 and offer potential targets for therapeutic intervention (39, 40).
The implications of our study for future research and clinical practice are significant. The identification of specific miRNAs that correlate with IL-6 levels in COVID-19 patients suggests their potential use as biomarkers for early diagnosis and risk stratification (41, 42). To confirm these results and investigate their potential clinical utility, more studies with larger sample sizes are required. Additionally, therapeutic interventions targeting these miRNAs could be developed to modulate the inflammatory response, potentially improving clinical outcomes in COVID-19 patients (25, 43-46).
Although significant correlations have been found between increased expression of specific miRNAs (miR-200b-3p, miR-34b-5p, miR-203a-3p, Let-7g-5p) and IL-6 levels in COVID-19 patients, several limitations should be acknowledged. First, the small sample size may limit the statistical power and hinder the applicability of findings across various patient demographics and disease severities. Second, even though the existing literature and our results indicate the potential for these miRNAs to impact inflammatory mediators via pathways such as NF-κB signaling, IFN responses, and the stabilization of IL-6 mRNA, our investigation did not experimentally confirm these mechanistic links. The lack of functional assays (such as luciferase reporter assays or the use of miRNA mimics or inhibitors in cell models) restricts our ability to establish causal relationships regarding the regulatory functions of the identified miRNAs.
Moreover, the study's cross-sectional approach doesn't allow for tracking how miRNA levels change over time as the disease progresses or in response to treatment. To address this, future research should focus on larger sample sizes, longer-term studies, and experiments that explore the biological mechanisms behind these miRNAs, including their direct targets and the signaling pathways they affect. Also, using methods that combine different types of biological data, like gene expression and protein levels, might offer a clearer picture of how miRNAs influence the immune system. In the long run, testing whether these miRNAs can predict disease outcomes or be used as treatments in large groups of patients could lead to new strategies for managing the harmful inflammation seen in severe cases of COVID-19.
5.1. Conclusions
In conclusion, our study underscores the importance of miR200b-3p, miR34b-5p, miR203a-3p, Let7g-5p, and IL-6 in the context of COVID-19. The significant relationships between these miRNAs and IL-6 levels highlight their potential as biomarkers and therapeutic targets, paving the way for advancements in the diagnosis, prognosis, and treatment of COVID-19.