1. Background
Haemophilus influenzae has been classified into two major groups, based on the presence of polysaccharide capsule; unencapsulated (nontypeable H. influenzae) (NTHi) and encapsulated (typeable H. influenzae) strains, in both of which the pe gene that is responsible for the production of adhesion protein, is present (1, 2). There are six types of encapsulated H. influenzae: a, b, c, d, e, and f (3). H. influenzae type b (Hib) has been recognized as the most virulent type. Bacteremia, pneumonia, and acute bacterial meningitis are diseases caused by Hib. In addition, otitis media, sinusitis in children, conjunctivitis and pneumonia are caused by NTHi (4-8). The initial adherence to the respiratory mucosa is the major factor for H. influenzae pathogenesis. When the bacteria are attached to the epithelium, colonization occurs in nasopharyngeal airway. As a result of impaired epithelium, an inflammation response and deep tissue penetration can occur (9). Usually, numerous surface-exposed proteins of H. influenzae including auto-transporters (ie, Hap, HMW1/HMW2, Hia, Hsf, P2, P5, D and E proteins) are involved in adherence to epithelial cells (10, 11).
Autotransporters are a superfamily of proteins which use type V secretion pathway for delivery to the surface of Gram-negative bacteria. In Gram-negative bacteria, navigations through inner membrane, periplasm and outer membrane are facilitated via seven secretion pathways including types I - VI and the chaperon-usher, which greatly vary in complexity, structural features and mechanisms of protein translocation. The mechanisms of type V secretion pathway are the simplest and most widely used mechanisms for delivery of proteins to the surface of Gram-negative bacteria (12). Autotransporters seems to contain all the required functional elements to promote their own secretion, including an amino-terminal signal peptide, a central passenger domain, and a channel-forming carboxyl terminus, which respectively can mediate translocation across the inner membrane, serve as a functional moiety, and facilitate passenger domain translocation across the outer membrane. However, recent discoveries on their structural and functional properties have challenged the perceived simplicity of autotransporter secretion (12). One of the autotransporters of H. influenzae is a 16 - 18 kDa (160 amino acids) surface lipoprotein called protein E (PE) (2, 11); it is a major pathogenicity factor of H. influenzae and is conserved in both NTHi and encapsulated H. influenzae (96.9 - 100%). Amino acids 84 - 108 that constitute the epithelial cell binding region are completely conserved. Recent studies showed that compared to wild-type strains, the pe-deficient isogenic NTHi mutants significantly reduce adherence to epithelial cell lines (9, 13).
2. Objectives
In this study, the pe gene was cloned and expressed for obtaining the recombinant protein as a candidate for vaccine design. The aim of this study was to provide a prokaryotic high-level expression system to produce PE which is a predictable candidate for vaccine.
3. Materials and Methods
3.1. Bacterial Strains and Vector
Nontypeable H. influenzae ATCC 49766 was obtained from American type culture collection and cultured at 37°C on chocolate agar and then in Luria-Bertani broth (Merck, Darmstadt, Germany), supplemented with V factor (NAD) (Mast Group Ltd., Liverpool, UK) and X factor (hemin-L-histidine) (Sigma-Aldrich, Germany) and 5% CO2 (14). Escherichia coli TOP10 was selected for both cloning and expression analysis of the recombinant protein. The E. coli cells containing recombinant plasmids were cultured aerobically at 37°C in Luria-Bertani broth, containing 50 μg/mL ampicillin (Sigma, Saint Louis, MO, USA). Cloning and expression vectors were TA vector (pTZ57R/T) and pBAD/gIII A plasmid (Invitrogen, California, USA), respectively.
3.2. Amplification of Protein E
Genomic DNA of NTHi ATCC 49766 strain was extracted using phenol-chloroform extraction method (15). To obtain the complete sequence of the pe gene (483 bp), amplification by polymerase chain reaction (PCR) was performed. Specific primers were designed; the forward primer (5’-CATGCCATGGGAAAAATTATTTTAACA-3’) was designed with an NcoI restriction enzyme site, whereas the reverse primer (5’-GATCTCTAGATGTTTTTTATCAACTGAAAATGC-3’) was designed with an XbaI site for insertion of the pe gene into the multiple cloning sites of the vector. Amplification of the DNA was performed using high fidelity DNA polymerase (Fermentas, USA). The 20-μL reaction mixture contained 0.5 pmol (1 μL) of each primer, 2 μL of 10x high fidelity buffer with MgCl2, 0.2 mM of each dNTP (1 μL), 2.5 U (0.5 μL) of high fidelity DNA polymerase, 100 ng genomic DNA (1 μL); the total volume was reached to 20 μL with double distilled water (DDW). Amplification was achieved using 35 cycles of denaturation at 94°C for 30 seconds, annealing at 57°C for 60 seconds, extension at 72°C for 90 seconds, and 10 minutes at 72°C for the final extension in a Techne thermocycler (Bibby Scientific Ltd., Staffordshire, UK). The PCR product was visualized on 1% agarose gel and was eluted from the gel and purified by gel electrophoresis with DNA molecular weight marker. Purification was then performed using the GeneJET purification kit (Thermo Fisher Scientific, USA).
3.3. Cloning and Expression of Protein E
The purified pe gene was ligated into cloning TA vector (pTZ57R/T) and was sequenced by a commercial facility using universal M13 primers. The TA-PE Recombinant vector was transformed into competent E. coli TOP10 cells. For attaining a positive transformant, colony-PCR with specific primers was performed. Next, the recombinant vector was extracted by QIAprep® Miniprep DNA purification system (Qiagen, Crawley, UK), and tested by restriction endonuclease enzymes; pe gene with an NcoI site at the 5’ end and an XbaI site at the 3’ end was achieved. The gene was subcloned into the NcoI-XbaI sites of pBAD/gIII A vector for expression of the recombinant protein. The pBAD/gIII A vector was digested with the restriction enzymes NcoI and XbaI and the pe gene was ligated into the NcoI-XbaI sites of the pBAD/gIII A vector, which provides six His sites at the N-terminus of the expressed recombinant protein. The pBAD/gIII A-PE recombinant vector was transformed into competent E. coli TOP10 cells. The transformants were chosen by colony PCR with specific primers and then the recombinant vector was extracted by QIAprep® Miniprep DNA purification system (Qiagen) and tested by restriction endonuclease digestion.
Nucleotide sequencing was performed by a commercial facility using forward and reverse primers of pBAD/gIII A (pBAD forward: ATGCCATAGCATTTTTATCCA, pBAD reverse: GATTTAATCTGTATCAGG). For the expression, the TOP10 cells containing a recombinant vector pBAD/gIII A-PE were cultured and grown in Luria-Bertani broth medium (MERCK, Germany) containing 50 μg/mL ampicillin, with shaking (250 rpm) at 37°C. For the induction, when optical density (OD) = 0.5 was obtained, arabinose in different concentrations of 20, 2, 0.2, 0.02, and 0.002 mmol/L were used to achieve the best concentration. The cells were incubated for extra four hours at 37°C. Afterwards, the induced cells (the pellet of TOP10 cells harboring PE) were obtained by centrifugation at 4500 g for 15 minutes at 4°C. The pellets were suspended in PBS1x and loading solution containing SDS, 2-mercaptoethanol (2ME), and loading buffer. The amounts of protein expression in each concentration were compared through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), using 15% polyacrylamide gel. The gel was then stained by standard coomassie brilliant blue G-250 for four hours and destained by 45% methanol and 10% acetic acid solution for four hours and the best concentration was obtained.
3.4. Purification of Recombinant Protein E
For purification of PE, affinity chromatography on a nickel-nitrilotriacetic acid (Ni-NTA) gel matrix (Qiagen, Crawley, United Kingdom) was performed under natural conditions. A pellet of TOP10 cells harboring PE was thawed on ice for 15 minutes and then suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH = 8). In the next step, lysozyme was added to 0.01 mg/mL and incubated on ice for 30 minutes. The solution was sonicated (five portions of 1-minute bursts at 300 W with a 10 second cooling period between each burst) and centrifuged for 30 minutes at 10000 g at 4°C. One milliliter of Ni-NTA was added to the pellet; it was then loaded in the column and rotated for one hour at 4°C. The column was washed with five volumes of washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH = 8) and subsequently with three volumes of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH = 8). Purification was performed using SDS-PAGE followed by coomassie blue staining. Protein concentrations were determined using Nanodrop analyzer (Bio-Rad, Hercules, CA, USA).
3.5. Anti-Protein E Antibody Production
Female New Zealand white rabbits were obtained from Pasteur institute of Tehran, Iran and immunized subcutaneously with 50 μg native NTHi purified PE in complete Freundʼs adjuvant (Sigma, Saint Louis, MO, USA). After 14 and 28 days, booster doses were injected in incomplete Freund’s adjuvant and the animals were exsanguinated 10 days after the last immunization. Serum was separated and stored at -20°C.
3.6. Western Blot Analysis
After running and separating the protein by SDS-PAGE, Western blot analysis with a 0.45-μm pore polyvinylidene difluoride (PVDF) membrane and anti-His antibody was carried out for identification of the recombinant protein.
4. Results
4.1. Amplification of Protein E
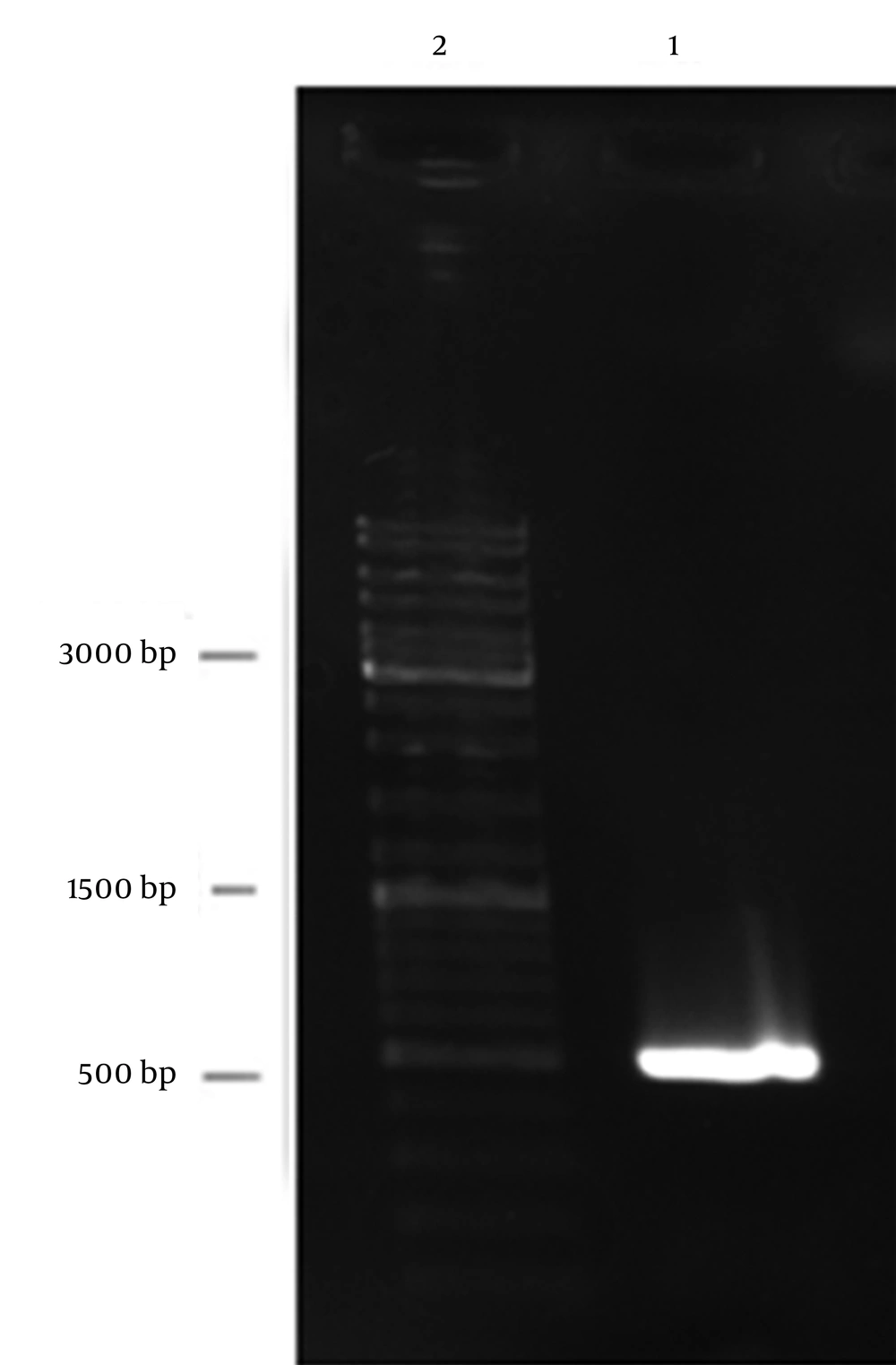
After amplification, gel electrophoresis with DNA molecular weight marker was carried out; the expected size of PE PCR product was approximately 483 bp (Figure 1).
4.2. Cloning of Protein E
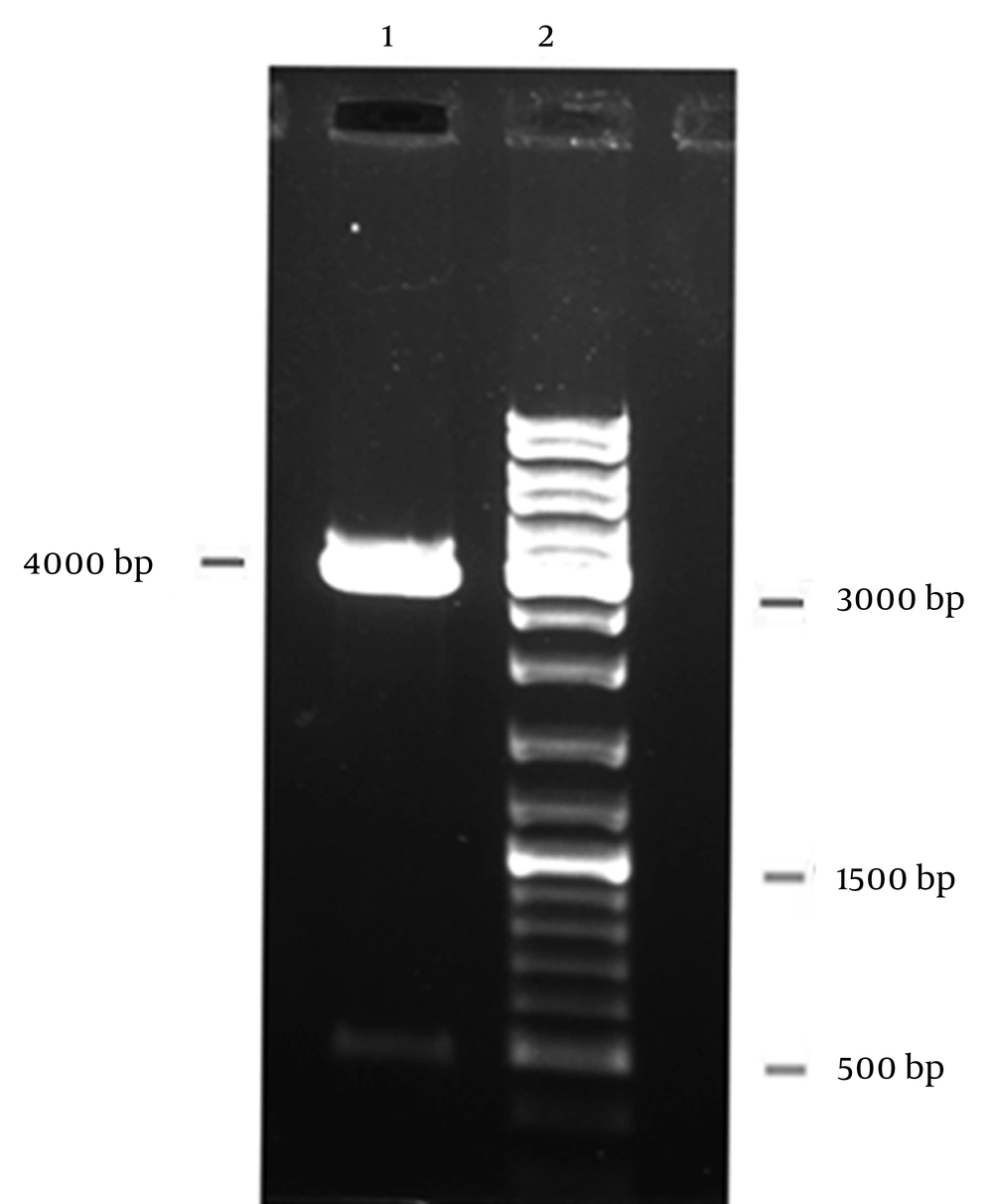
Colony-PCR with specific primers confirmed a positive transformant and double-digestion by NcoI and XbaI restriction enzymes confirmed the recombinant vector pBAD/gIII A-PE (Figure 2). For identification of the pe gene structure, nucleotide sequencing was carried out and nucleotide sequence comparison between the cloned pe gene and the reference sequences of pe gene in GenBank showed 99% similarity. The gene was submitted in GenBank with accession number KF366252.
4.3. Expression of Protein E
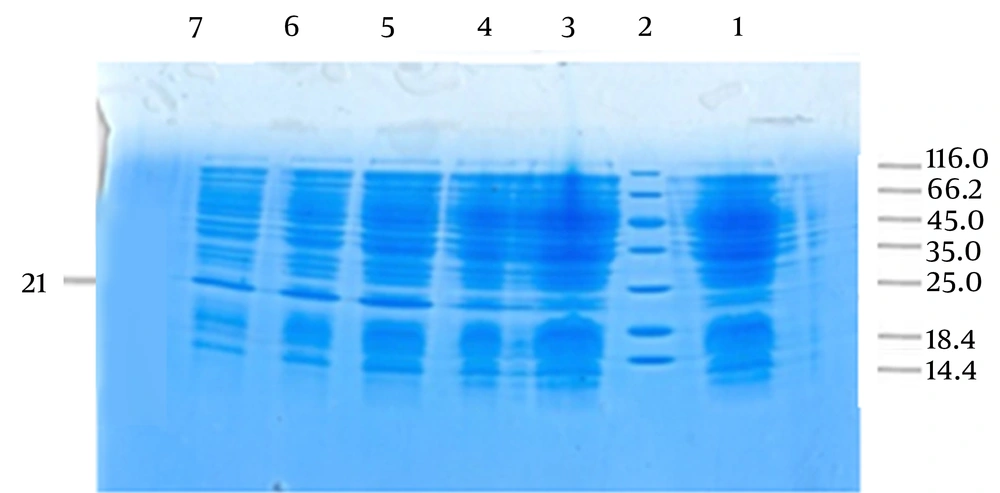
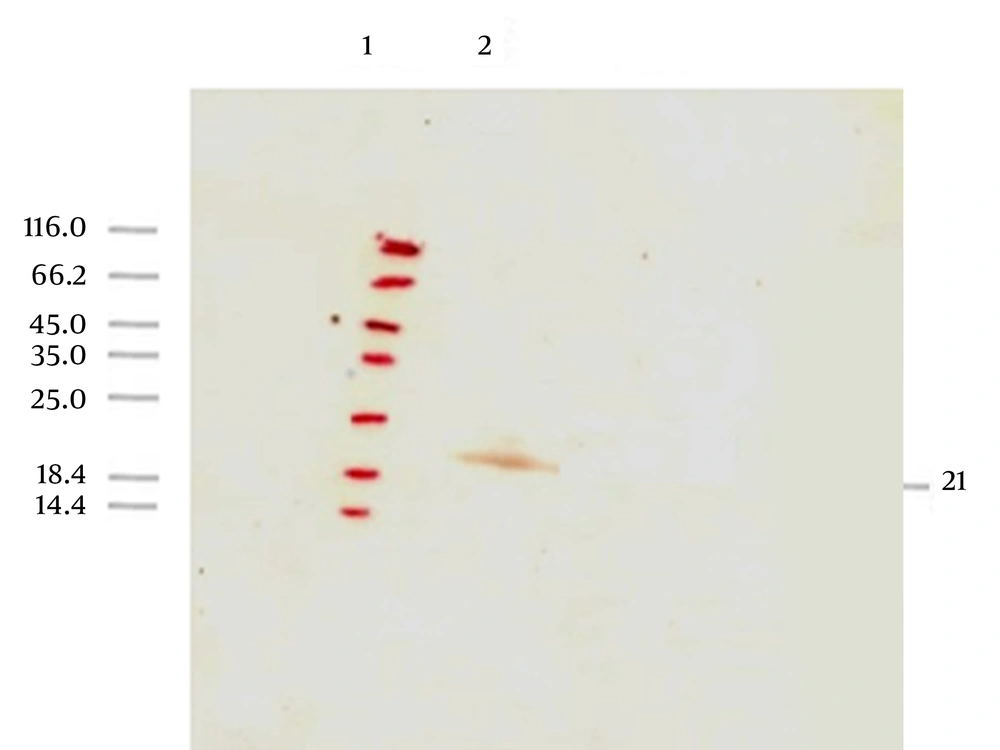
SDS-PAGE analysis showed a sharper band than other bands between the 18.4 and 25 kDa positions (approximately 21 kDa), which indicated the position of PE. PE is an 18 kDa protein and the observed difference in size was due to the addition of pBAD/gIII A vector signal peptide. After performing SDS-PAGE using 18% polyacrylamide gel, the results showed that the best induction was 0.002 mmol/L arabinose for four hours (Figure 3).
4.4. Purification of Protein E
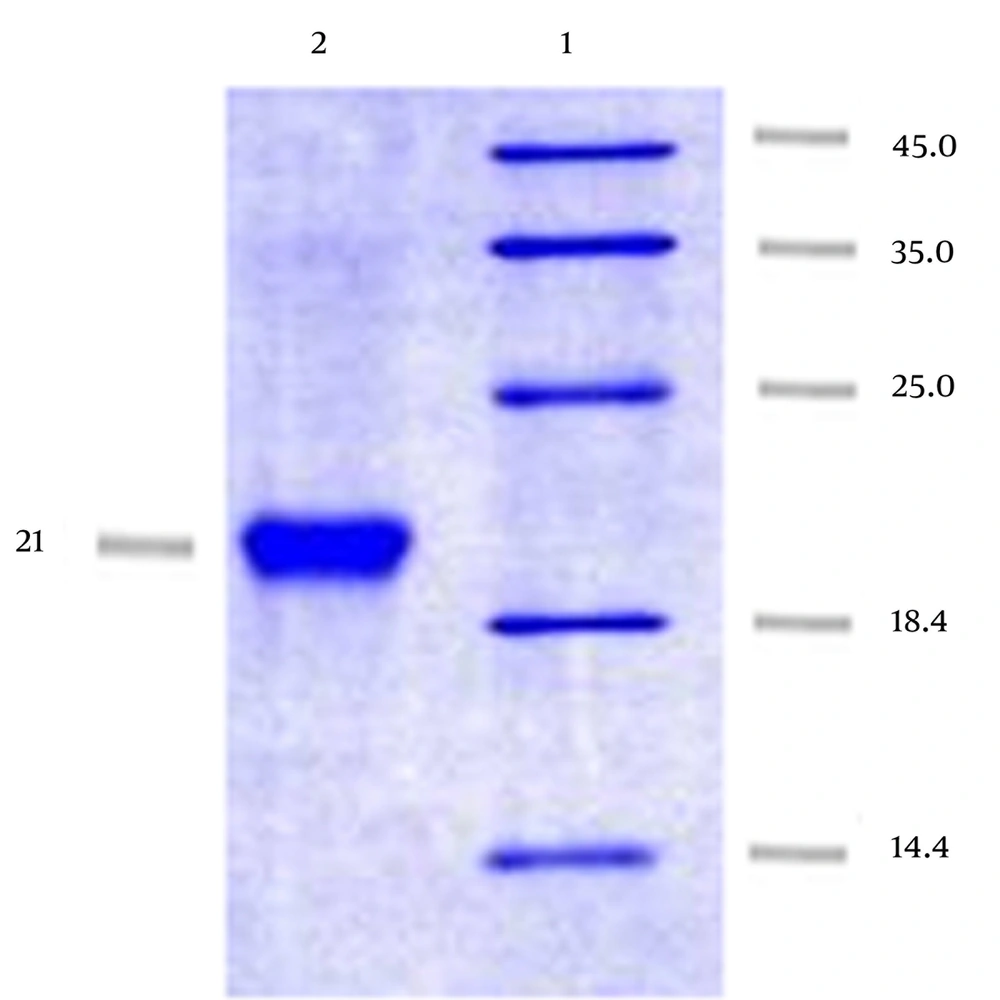
We successfully obtained PE using affinity chromatography on Ni-NTA gel matrix under natural conditions with up to 7 mg/mL concentration (Figure 4).
4.5. Western Blot Analysis
A band between 18.4 and 25 kDa (approximately 21 kDa) was observed after performing SDS-PAGE and then Western blot with anti-His antibody which revealed the identification and evaluation of the expressed protein antigenicity (Figure 5).
5. Discussion
NTHi has different surface proteins, many of which are not conserved. In recent years, some NTHi outer membrane proteins have been studied and reported as vaccine candidates such as P1/P2/P6 (16 kDa) and protein D (45 kDa) (10, 16), high molecular weight proteins (HMW-1 and HMW-2) (125 kDa) (17, 18), the Hia homologue Hsf protein (approximately 245 kDa) (19), Hap protein (110 kDa) (10), and PE (16-18 kDa) (11). PE is a small surface lipoprotein comprising only 160 amino acids, which could act as a tight adherence of H. influenzae to different types of the epithelial cells (11). Based on Singh et al. studies in 2010, PE has a conserved central core domain which constitutes a potential epithelial binding region (84 - 108 amino acids); so, it can be considered as a possible vaccine candidate (4). Hallstrom et al. in 2009 showed that this protein was conserved in both NTHi and encapsulated H. influenzae (96.9–100%) (13). The initial adherence to the respiratory mucosa is the major virulence factor of H. influenzae (9, 13).
In this study, we used a prokaryotic high level expression recombinant system (pBAD gIII A-PE-E. coli TOP10) which can produce recombinant PE. We performed cloning, expression and purification of PE by affinity chromatography on Ni-NTA gel matrix; His-tag facilitates purification of recombinant protein by Ni+2-sepharose resin. The pe gene was cloned into TA vector (pTZ57R/T) and then sub-cloned in pBAD/gIII A vector and transformed into competent E. coli TOP10. Due to the presence of high similarity among PE from NTHi ATCC 49766 and other NTHi strains in GenBank, it seems that this gene could be a vaccine candidate against NTHi. To overexpress, the recombinant bacteria were grown in broth medium containing arabinose and then the recombinant protein was purified using metal affinity chromatography (Ni-NTA agarose).
Singh et al. (2) expressed PE as inclusion bodies in E. coli BL21 (DE3) with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) as inducer and purified PE by Q-sepharose fast flow anion-exchange column. They concentrated the recombinant PE up to 5 mg/mL.
In this study, harboring vector of the pe gene was transformed into competent E. coli TOP10 cells and expressed with 0.002 mM arabinose. We obtained PE protein using affinity chromatography on Ni-NTA gel matrix under natural conditions with up to 7 mg/mL concentration. Based on the comparison of our pe gene with corresponding sequences of original genes, the nucleotide sequence homology of the cloned pe gene was 99%. GenBank accession number for our nucleotide sequence is KF366252. Due to observation of highly conserved amino acids among the PE from NTHi ATCC 49766 and other NTHi strains in GenBank, it seems that the protein is a universal antigen and antibody response against it could protect against different NTHi strains. Therefore, further studies for preparation and immunobiological evaluation of two- or three-valent complexes of NTHi PE with other bacterial conserved proteins for protecting against both NTHi and other bacteria can be targeted in vaccine studies. The recombinant protein was highly expressed in E. coli and further studies to evaluate its immunogenicity are underway; its production and extraction are crucial step for further in vivo studies.