1. Background
For the first time in 1949, it was reported that crystalline glycolipid isolated from Pseudomonas aeruginosa showed antibiotic activity against tuberculosis in mice (1). At that time, there was little knowledge about biosurfactants and enormous capabilities of these compounds. Several decades later, in a world based on green technology as a foundation of the search for sustainable development, biological compounds were being recognized increasingly as an alternative to their synthetic counterparts because of characteristics such as mild process conditions, low toxicity, high biodegradability, and environmental compatibility (2-8). Biological surfactants (biosurfactants) are an example of such compounds.
Biosurfactants are produced by diverse microorganisms to fulfill various natural functions. They exhibit a broad spectrum of chemical structures such as glycolipids, lipopeptides and lipoproteins, lipopolysaccharides, phospholipids, fatty acids, and polymeric lipids (9, 10). Therefore, it is reasonable to expect various properties and physiological functions for biosurfactants such as reducing surface and interfacial tensions as well as increasing surface area and bioavailability of hydrophobic water-insoluble compounds, heavy metal binding, bacterial pathogenesis, quorum sensing, and biofilm formation (11). Due to their ability to dissolve or disperse hydrophobic compounds in water, biosurfactants have prompted in food, pharmaceutical, and cosmetic applications as well as in crop protection, environmental industries, energy-saving technologies, and so on (2-5, 7, 8, 12, 13).
Some of interesting features of biosurfactants have led to a wide range of potential applications in the medical field. They are useful as antibacterial, antifungal, and antiviral agents (14). Particularly, these products are highly valued for their antimicrobial activities and lack of toxicity (15). The antimicrobial activity of several biosurfactants has been reported in the literature for various applications (16).
Different types of biosurfactants have shown antimicrobial actions against bacteria, fungi, algae, and viruses. The lipopeptide iturin from Bacillus subtilis showed potent antifungal activity (17). Inactivation of enveloped viruses such as herpes virus and retrovirus was observed by 80 microM of surfactin (18). Rhamnolipids inhibited the growth of harmful bloom algae species, Heterosigma akashivo, and Prorocentrum dentatum at concentrations ranging from 0.4 to 10.0 mg/l. A rhamnolipid mixture obtained from P. aeruginosa AT10 showed bacterial inhibiting activity against Escherichia coli, Micrococcus luteus, Alcaligenes faecalis (32 mg/mL), Serratia marcescens, Mycobacterium phlei (16 mg/mL) and Staphylococcus epidermidis (8 mg/mL), and excellent antifungal properties against Aspergillus niger (16 mg/mL), Chaetomium globosum, Penicillium chrysogenum, Aureobasidium pullulans (32 mg/mL), and phytopathogenic Botrytis cinerea and Rhizoctonia solani (18 mg/mL) (19). Sophorolipids and rhamnolipids were found to be effective antifungal agents against plant and seed pathogenic fungi. The mannosylerythritol lipid (MEL), a glycolipid surfactant from Candida antarctica, has demonstrated antimicrobial activity particularly against Gram-positive bacteria (20).
Biosurfactants appears to be great potential and suitable alternatives to synthetic medicines and antimicrobial agents and may be used as safe and effective therapeutic agents (3, 11, 14, 21). Glycolipids are the most promising among biosurfactants due to high productivity from renewable resources and versatile biochemical properties (2-8). There have been many previous studies on antimicrobial activities of rhamnolipids produced by P. aeruginosa strains and they were known to exhibit this feature activity (19, 22-24).
2. Objectives
This study specifically aimed to compare antibacterial effects of biosurfactants produced by different P.aeruginosa strains isolated from different sources when grown on different carbon sources. MR01 and MASH1 strains were isolated from two oil excavation areas in south of Iran.
3. Materaials and Methods
3.1. Biosurfactants and Test Microorganisms
MR01 biosurfactant produced by P. aeruginosa MR01 with GenBank accession number of EU795302 in M1 medium using glucose as sole carbon source was purified as described in previous work (25).
MASH1 biosurfactant produced by MASH1 (26) using soybean oil as sole carbon source was cultivated in SOM medium composed of (g/L): NaNO3 3.0, KH2PO4 0.25, MgSO4.7H2O 0.25, yeast extract 1.0 and soybean oil 10. Seed cultures were prepared using a seed culture medium containing (g/L): glucose 40, NaNO3 3.0, KH2PO4 0.25, MgSO4.7H2O 0.25 and yeast extract 1.0 by overnight incubation on rotary shaker at 200 rpm and 30°C. Seed cultures were inoculated (2% v/v) in 250 ml flasks containing 50 ml of production medium followed by incubation at 200 rpm and 30 °C for 6 days. Crude biosurfactant was obtained using an acid precipitation and solvent extraction method (27, 28) . At the end of incubation, cells were removed from the culture broth by centrifugation at 10000 × g for 15 min at 4 ˚C. The cell free supernatant was acidified with 6 N HCl to pH 2 and stored overnight at 4 ˚C to enhance precipitation of biosurfactant. Then it was centrifuged (18000 × g, 30 min, 4 ˚C) and the resultant precipitate was extracted several times with ethyl acetate at room temperature. The solvent was completely evaporated under vacuum. The crude biosurfactant was obtained as a viscose brown-colored material (26).
The microorganisms used in this study included laboratory control strains of Streptococcus pneumonia (clinical sample), S. epidermidis (ATCC 12228), S. aureus (ATCC 29213), B. subtilis (ATCC 6051), B. cereus (PTCC1247), Enterococcus faecalis (ATCC 29212), E. faecalis (clinical sample) and Micrococcus luteus, (clinical sample) as Gram-positive bacteria. Salmonella typhi (ATCC19430), Klebsiella pneumonia (clinical sample), Klebsiella pneumonia (ATCC 7881), E. coli (clinical sample), E. coli (ATCC 25922), E. coli K12 (clinical sample) and P. aeruginosa (ATCC 27853) as Gram-negative bacteria were obtained from Department of Bacteriology, Pasteur Institute of Iran, Tehran, Iran.
Evaluated fungal strains were A. niger ATCC 14604, A. pullulans ATCC 9348, C. globosum ATCC 6205, P. chrysogeum CECT 2802, and P. funiculosum CECE 2914.
3.2. Evaluation of Antibacterial Effects by Disc Diffusion Method
The antibacterial effects of MR01 and MASH1 biosurfactant samples were assessed separately in every 3 concentrations of 10, 20, and 30 mg/ml on E. coli ATCC 25922, P. aeruginosa ATCC 27853, S. aureus ATCC 29213, S. epidermidis ATCC 12228, and B. subtilis ATCC 6051.
The Kirby-Bauer disc diffusion susceptibility test (28) performed in accordance to Clinical and Laboratory Standards Institute (CLSI) 6mm discs containing 30μl of the different dilutions of two biosurfactant samples were applied to the surface of medium. The plates were incubated at 35°C aerobically (no CO2) for 16-18 h. Then each plate was examined and diameters of the zones of complete inhibition, together with diameter of the disk were measured.
3.3. Evaluation of Antibacterial Effects Using Minimal Inhibition Concentration (MIC) Method
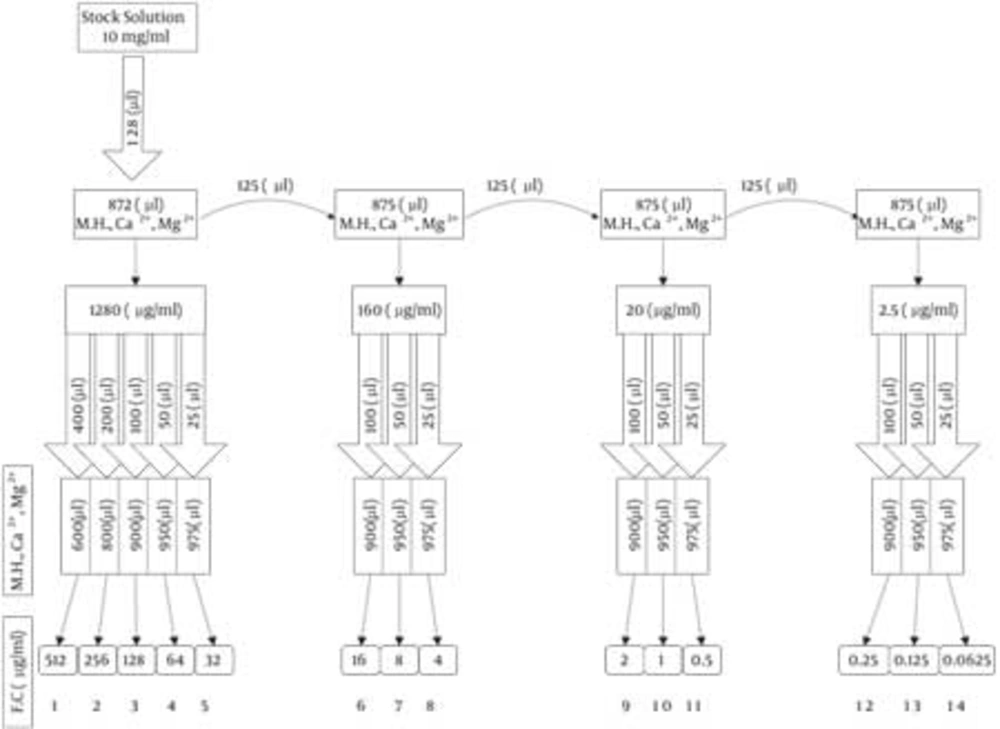
Antimicrobial activity was determined on the basis of minimal inhibition concentration (MIC) values, defined as the lowest concentration of antimicrobial agent needed to inhibit the development of visible growth after incubation for the required time. A standard two-fold serial rhamnolipid dilution technique was applied to measure antimicrobial activity (29). Briefly, a 10 mg/ml stock solution of MR01 and MASH1 biosurfactants in Muller Hinton broth containing Ca2+, Mg2+ was diluted twofold to concentrations ranging from 0.25 to 512 μg/mL (Figure 1).
Antibacterial activity was determined on 15 bacterial suspensions (Table 1 and Table 2).
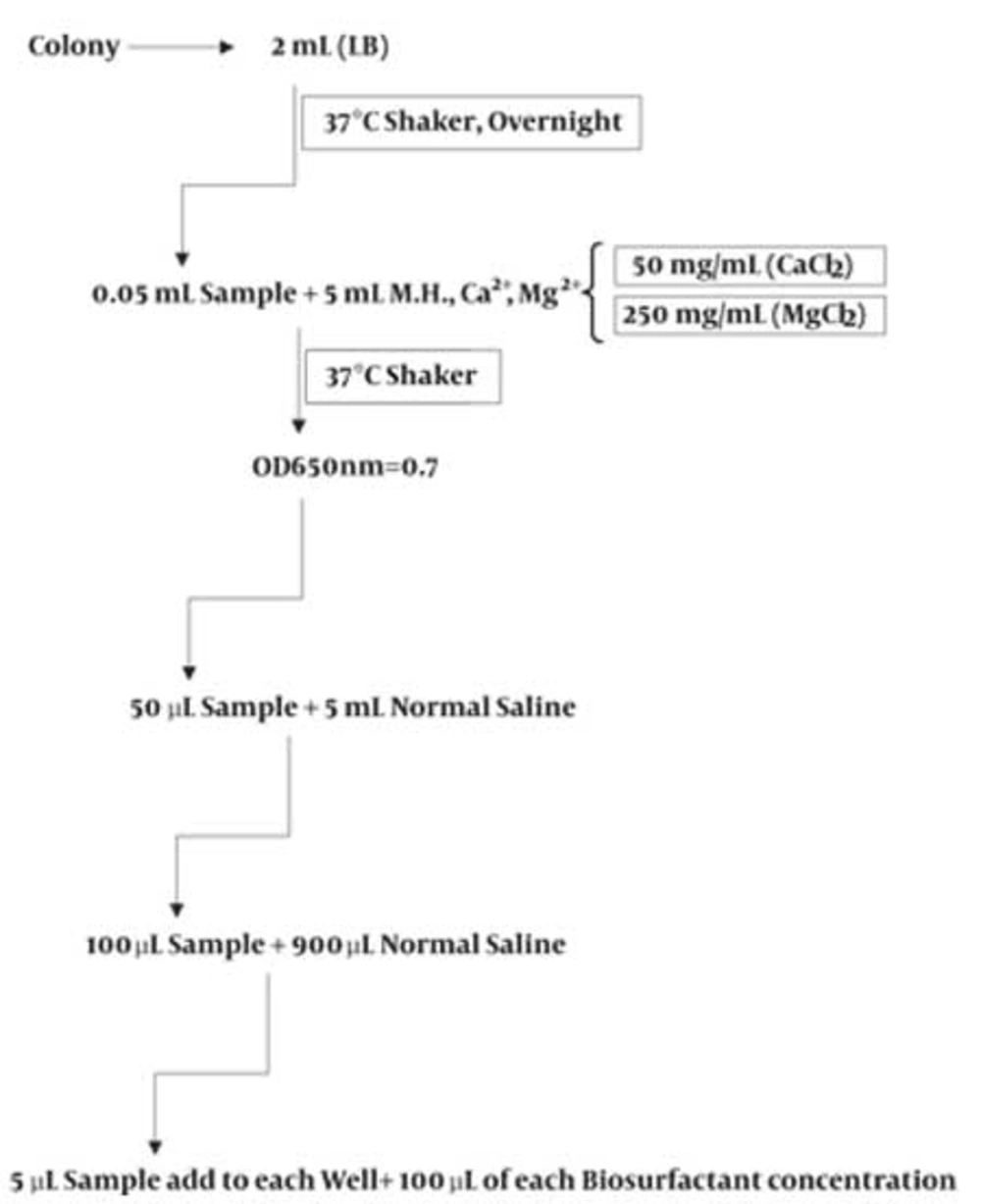
For each strain tested, one colony with typical morphology was selected from an agar plate that had been incubated overnight and inoculated into the Muller Hinton broth containing Ca2+, Mg2+ which was incubated in Muller-Hinton broth at 37°C for 12 h. The cell density of the cultures was adjusted to approximately 1×108 cfu/mL, equivalent to an absorbance reading at 650 nm of 0.7 and then diluted in normal saline as showed in Figure 2.
| Bacteria | Code | Zone Inhibition Diameters (nearest whole mm) | ||
|---|---|---|---|---|
| Biosurfactant Concentration, mg/mL, MR01, MASH1 | ||||
| 10 | 20 | 30 | ||
| Gram positive | ||||
| Staphylococcus aureus | ATCC 29213 | 14 ± 1, 20 ± 2 | 15 ± 2, 20 ± 2 | 15, 20 ± 2 |
| Staphylococcus epidermidis | ATCC 12228 | 15 ± 2, 20 ± 3 | 15 ± 2, 20 ± 2 | 15 ± 2, 20 ± 3 |
| Bacillus cereus | ATCC 6051 | 30 ± 3, 30 ± 2 | 30 ± 3, 30 ± 2 | 30 ± 3, 30 ± 2 |
| Gram negative | ||||
| Escherichia coli | ATCC 25922 | 0, 0 | 0, 0 | 0, 0 |
| Pseudomonas aeruginosa | ATCC 27853 | 0, 0 | 0, 0 | 0, 0 |
Disc Diffusion Susceptibility of Biosurfactant Produced by MR01 and MASH1 Strains Using Glucose and Soybean Oil as Sole Carbon Source, Respectively.
| Bacteria | Code | MIC(μg/mL) | |
|---|---|---|---|
| MR01 | MASH1 | ||
| Gram positive | |||
| Streptococcus pneumonia | (clinical sample) | 128 | 128 |
| Staphylococcus epidermidis | (clinical sample) | 128 | 64 |
| Staphylococcus aureus | ATCC 29213 | 128 | 512 |
| Bacillus subtilis | ATCC 6051 | 128 | 128 |
| Bacillus cereus | PTCC1247 | 128 | 64 |
| Enterococcus faecalis | ATCC 29212 | > 512 | 64 |
| Enterococcus faecalis | (clinical sample) | > 512 | 64 |
| Micrococcus luteus | 32 | 64 | |
| Gram negative | |||
| Salmonella typhi | ATCC 19430 | > 512 | > 512 |
| Klebsiella pneumonia | (clinical sample) | > 512 | > 512 |
| Klebsiella pneumonia | ATCC 7881 | > 512 | > 512 |
| Escherichia coli | (clinical sample) | > 512 | > 512 |
| Escherichia coli | ATCC 25922 | > 512 | > 512 |
| Escherichia coli K12 | > 512 | > 512 | |
| Pseudomonas aeruginosa | ATCC 27853 | > 512 | > 512 |
Minimum Inhibitory Concentration Results
3.4. Evaluation of Antifungal Activity Using Growth Inhibition Method
Antimicrobial activity of MR01 and MASH1 biosurfactants was detected on the basis of minimal inhibition concentration (MIC) values, defined as the lowest concentration of antimicrobial agent needed to inhibit the development of visible growth after incubation for the required time. Antimicrobial activity against fungi was accomplished on solid medium Sabouraud agar plates which were incubated for 72 h at 25°C (30).
4. Results
Disc diffusion data for susceptibility of test strains against MR01 and MASH1 biosurfactant has been represented in Table 1. Table 2 shows the comparison of MIC results of biosurfactants produced by MR01 and MASH1 strains against different Gram positive and Gram negative bacteria. The results for inhibitory activities of MR01 and MASH1 biosurfactants against several reference fungal strains have been shown in Table 3.
| Fungi | Code | MIC, µg/mL | |
|---|---|---|---|
| MR01 | MASH1 | ||
| Aspergillus niger | ATCC 14604 | 64 | 128 |
| Aureobasidium pullulans | ATCC 9348 | 128 | 128 |
| Chaetonium globosum | ATCC 6205 | 64 | 32 |
| Penicillium chrysogeum | CECT 2802 | > 256 | 128 |
| Penicillium funiculosum | CECE 2914 | 32 | 32 |
Minimal Inhibitory Concentration (MIC) of MR01 and MASH1 Biosurfactants Against Selected Fungi.
5. Discussion
Preliminary disc diffusion data summarized in Table 1, showed that all examined Gram positive bacteria (i.e. S. aureus ATCC 29213, S. epidermidis ATCC 12228 and B. cereus ATCC 6051) were inhibited by biosurfactants produced by both MR01 and MASH1 strains. MASH1Growth of S. aureus ATCC 29213 and S. epidermidis ATCC 12228 were inhibited by MASH1 biosurfactant more than that by MR01 biosurfactant (exhibiting zone inhibition diameters 20 ± 2 and 15 ± 2 mm, respectively), While the inhibitory effects of both biosurfactants for B. cereus ATCC 6051 were similar. In this experiment both biosurfactants in different concentrations exhibited good antibacterial effects on Gram positive bacteria. None of examined Gram negative bacteria were affected by the MR01 and MASH1 biosurfactants produced from different carbon sources (Table 1).
Regarding Table 2, none of biosurfactants affected the growth of Gram negative bacteria which confirmed the results of our preliminary tests based on disc diffusion method. On the other hand, MR01 and MASH1 biosurfactants showed different antimicrobial activity behaviors on Gram positive bacteria. The MIC values of MR01 biosurfactant for S. epidermidis ATCC 12228, B. cereus PTCC1247, E. faecalis ATCC 29212, and E. faecalis (clinical sample) were higher than that of MASH1 biosurfactant. Growth of both E. faecalis strains inhibited at the presence of MASH1 biosurfactant at approximately low concentration (64 μg/mL), while MR01 biosurfactant couldn’t inhibit the growth of those bacteria at concentration below 512 μg/mL. MIC values of MR01 for S. aureus ATCC 29213 and M. luteus (clinical sample) were lower than that of MASH1 biosurfactant and respective values for Streptococcus pneumoniae (clinical sample) and B. subtilis ATCC 6051 were equal for both biosurfactants (Table 2). The results of the second method (MIC) were more delicate than the former because in the latter, the sensitivity was high and number of tested bacteria was increased.
Gram positive bacteria studied in this work were more susceptible to both biosurfactant samples than Gram negative ones. It means that examined biosurfactants were more active against Gram-positive bacteria than against Gram-negative ones (Table 2). This is an important finding as there is still little information about antimicrobial activity of surfactants.
Several structural rhamnolipid congeners produced by different strains of P. aeruginosa have been identified so far (22, 30-32).The type of rhamnolipid produced depends on the bacterial strain, the carbon source used, and the process strategy (24, 33, 34). Rhamnolipids are formed by one or two rhamnose molecules linked to one or two fatty acids of saturated or unsaturated alkyl chain between C8 and C12 (35). New analytical methods of high-performance liquid chromatography and mass spectrometry (HPLC-MS) have identified numerous congeners which contain one or two rhamnose molecules and one or two residues of 3-hydroxydecanoic acid groups (30-32, 34, 36, 37). Even 3-hydroxydodecenoic and 3-hydroxytetradecenoic acid residues have been reported (34, 36).
Similar results for being more effective on Gram positive bacteria than that on Gram negative ones were previously reported for glycolipid biosurfactant of MEL by Kitamoto et al. (20). and rhamnolipid biosurfactant of P. aeruginosa AT10 by Haba et al. (37). Relying on therapeutic applications of some biosurfactants as antibiotics and antifungal or antiviral compounds, some of researchers studied on several surfactant-resistant strains and isolated sensitive strains of bacteria by artificial or spontaneous mutagenesis and concerned this phenomenon to possess altered structures and functions in their cell membranes (38, 39).
Yilmaz and Sidal reported that the antimicrobial activity against Gram positive bacteria was more potent than against Gram negative bacteria. They supposed that mechanism of antimicrobial action of rhamnolipid biosurfactant regards to the fact that biosurfactants may disturb membrane structure through interaction with phospholipids as well as membrane proteins. Biological function of biosurfactants has not been completely understood yet; e.g., these substances, when excreted into the medium, emulsify hydrocarbons, and when located in cell wall structure , facilitate the penetration of hydrocarbons to periplasmic space (40). Onbasli et al. studied relationship between antimicrobial activity and rhamnolipid production and explained observed findings by hardly permeable characteristic of Gram negative bacteria membranes to hydrophobic and amphipathic molecules . It is well-known that Gram positive and Gram negative bacteria have different cell wall chemical structures (41).
According to disc diffusion and MIC results, both biosurfactants had no significant effects on Gram-negative bacteria; this finding was rather different with previous results reported by Haba et al. about effects of rhamnolipids produced by P. aeruginosa 47T2 NCBIM 40044 on Gram negative bacteria (22). The variation observed in results may be attributed to the differences in homologues composition of the rhamnolipid mixtures evaluated. When Haba et al. compared antimicrobial activities of two rhamnolipid mixtures obtained from different P. aeruginosa strains, they observed that P. aeruginosa AT10 rhamnolipids were most effective against fungi, and P. aeruginosa 47T2 rhamnolipids was more efficient against bacteria. The effects observed were attributed to differences in homologue composition of both rhamnolipid mixtures (22).
Owing to their intrinsic properties, surface-active compounds interfere with cell surfaces and disrupt microbial membranes. Despite that there are many studies in relation to antimicrobial potency of rhamnolipids (19, 22-24, 41, 42), still there is little information about antimicrobial action mechanisms of rhamnolipid biosurfactants, nature and limitations of antibacterial spectrum of natural rhamnolipid congeners, and contribution of each homologous species in final activity of the product (24). To our best knowledge, there are few reports in the literature on the physicochemical characterization and biological activity of rhamnolipid mixtures to be used as a single product. The properties of such a product depend on the type and proportion of homologous (19).
In our another study, HPLC-ES-MS in negative mode was used to identify rhamnolipid congeners produced by P. aeruginosa MR01. By performing this technique, up to 17 different rhamnolipid congeners were recognized in MR01 biosurfactant mixture when grown on glucose in M1 medium using glucose as sole carbon source (43). P. aeruginosa MR01, grown in M1 medium, produced a mixture of rhamnolipids with capability of surface tension reduction to 27 mN/m. P. aeruginosa MR01 produced several biosurfactants. MASH1 strain produced up to 6 different rhamnolipid congeners using soybean oil as sole carbon source (data not published). The different observed antibacterial effects against Gram positive bacteria for MR01 and MASH1 may be related to the differences in the culture medium composition, isolation source, carbon source, etc. MR01 was isolated from crude oil and produced biosurfactant using glucose as sole carbon source on M1 minimal salt medium at 37ºC, while MASH1 was isolated from other crude oil reservoir (26) andproduced biosurfactant using soybean oil as sole carbon source at 30ºC. These conditions may change the structure of biosurfactant produced by different strains and lead to different behavior of antibacterial properties of biosurfactants.
Regarding data shown in Table 3, high activity against C. globosum and P. funiculosum was observed by two MR01 and MASH1 biosurfactants but none of them was found to be effective on A. pullulans and P. chrysogeum. Indeed, MR01 and MASH1 biosurfactants exhibited different inhibitory behaviors against A. niger. MIC values obtained for MR01 and MASH1 biosurfactants against examined aforementioned fungi were slightly different with which reported by others (19, 22-23). It is supposed that the difference might be attributed to differences in homologues composition of rhamnolipid mixtures evaluated.