1. Background
Although the world’s share of camel milk in comparison with other domesticated animals is very low yet the milk produced by camels in drought areas can be a valuable food source for the human population (1). Iran is a country with vast arid regions. Under such conditions, the only livestock, which can successfully survive, is camel. However, camel milk has no access to the market and due to its poor keeping quality, it cannot be used as fresh and goes to waste. During the peak production season, it can be saved and effectively utilized through conversion to yoghurt and drinking yoghurt using a starter culture of Lactic Acid Bacteria (LAB). In fermented products from milk, LAB as starter culture play an important role in the process of fermentation. The microbiological quality of milk and milk products is influenced by the initial flora of raw milk (2). It has been recognized that LAB are capable of producing inhibitory substances such as organic acid, hydrogen peroxide, bacteriocin and diacetyl (3). Natural antimicrobials produced by microbes, especially LAB, have been widely used as chemotherapeutic agents that can control the growth of microbial pathogens (4).
One of the most significant groups of probiotic organisms is LAB. Probiotics are beneficial bacteria that favorably alter the intestinal microflora balance, inhibit the growth of pathogenic bacteria, promote digestion, boost immune function and increase resistance to infection. Other physiological benefits of probiotics include removal of carcinogens, lowering of cholesterol and immunostimulants, lowering the effect of allergies, alleviation of lactose intolerance. They must survive exposure to harsh conditions imposed by gastric acids (pH = 3), bile and digestive enzymes (0.4% bile salts) (5). A good tool for molecular identification of LAB is restriction analysis of 16S–23S rRNA intergenic spacer regions (6).
2. Objectives
The objectives of the present research were the isolation and molecular identification of LAB from camel milk and evaluation of their probiotic properties.
3. Materials and Methods
3.1. Isolation of Lactic Acid Bacteria
A total of ten samples of camel milk were collected from the Golestan province of Iran under aseptic conditions and appropriate dilutions of samples were plated onto four groups of plates including de Man Rogosa (MRS) agar and vancomycin, MRS agar alone, M17 agar, and Kanamycin Aesculin Azide (KAA) agar (Merck, Germany). Plates were incubated at 20°C, 37°C, 45°C for 48 hours under anaerobic conditions (using anaerocult® A gas packs, Merck, Germany). Colonies with different morphologies were counted and purified by re-streaking on the same medium. Gram staining, catalase test and microscopic observations were performed for preliminary screening of LAB. The isolated LAB strains were analyzed by gas production, 6.5% NaCl tolerance, growth at 10°C, 45°C, pH 4.4 and pH 9 and 6. The strains were identified at genus level (7).
3.2. Molecular Identification of Lactic Acid Bacteria Species
3.2.1. DNA Extraction
For DNA extraction, single colonies were resuspended in 50 µL of sterile deionized water. Next, 50 µL of chloroform/isoamyl alcohol (24:1) was added to the suspensions, and after vortexing, the mixture was centrifuged at 16 000 g for five minutes at 4°C. Then, 5 µL of the upper aqueous phase was used as a source of DNA template for the PCR reaction (8).
3.2.2. Amplification of the Internally Transcribed Spacer (ITS) Region and Analysis of the Amplified Ribosomal DNA
The primers used for the amplification of the ITS region between the 16S and 23S rRNA genes were B27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and L1R (5′-CAAGGCATCCACCGT-3′) (Bioneer, Korea) (9). The Polymerase Chain Reaction (PCR) was carried out by mixing 5 µL of each extracted DNA with 25 µL of 2X PCR Master kit (composition of 1X solution: 0.5 M Tris-HCl, 1.5 mM MgCl2 – 200 µM dATP, 200 µM dCTP, 200 µM dGTP and 200 µM dTTP and 0.04 Units/ul Taq) (SinaClon, Iran), 1 µL Oligo forward (10 picomole/µL), 1 µL Oligo reverse (10 picomole/µL) and 18 µL Sterile deionized water.
The amplification was achieved by 40 PCR cycles using the following procedure: first denaturation was performed at 94°C for five minutes, then 40 cycles at 94°C for one minute, 42°C for one minute and finally 72°C for one minute. After a final extension at 72°C for 10 minutes, the tubes were cooled to 4°C (10). The 16S rRNA- ITS gene PCR product from each LAB isolate was cut with restriction endonuclease enzyme: Taql and Hhal (Fermentas, USA), following the manufacturer’s instructions. The reaction mixture was then incubated at 65°C and 37°C for Taq I and Hhal, respectively for 1.5 hour. The Amplified Ribosomal DNA Restriction Analysis (ARDRA)profiles were examined using 1.5% (w/v) agarose gels in 0.5X Tris/Borate/Ethylenediaminetetraacetic acid (TBE) buffer at 75 V for 90 minutes with a DNA ladder (GeneRuler™ 100 bp) (Fermentas, USA).
3.2.3 Amplification of 16S rDNA Genes
Representative isolates of different ARDRA profiles were amplified by PCR. The primers used for the amplification of 16S rDNA genes were B27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and U1492R (5′-ACGTGGTTTGAAGAGATTTTCG-3′) (Bioneer, Korea). The PCR was carried out by mixing 5 µL of each extracted DNA with 25 µL of 2X PCR Master kit (SinaClon, Iran), 1 µL Oligo forward (10 picomole/µL), 1 µL Oligo reverse (10 picomole/µL) and 18 µL sterile deionized water. The amplification was achieved by 40 PCR cycles using the following procedure: first denaturation at 94°C for five minutes, then 40 cycles at 94°C for one minute, 56°C for one minute and 72°C for one minute. After a final extension at 72°C for 10 minutes, the tubes were cooled to 4°C (10). Representative amplicons of the different ARDRA profiles were purified and sequenced by Macrogen Sequencing Service (Korea).
3.3. Antimicrobial Activity Evaluation of Lactic Acid Bacteria
Pediococcus pentosaceus subsp. aureus ATCC 25923, Escherichia coli O157: H7 and Bacillus cereus ATCC 10876, were used as indicator strains. These were grown in Nutrient Agar (NA) at 37°C. Antimicrobial activity of LAB against E. coli O157 and B. cereus was determined by the agar well diffusion assay and against S. aureus subsp. aureus by disc diffusion assay. Isolates were grown in MRS and M17 broth (Merck, Germany) at 37°C for 24 hours. Each indicator bacteria was grown in nutrient broth (Merck, Germany) at 37°C for 24 hours. Furthermore, 100 μL of this broth culture of pathogenic bacteria was cultured on Muller-Hinton Agar (MHA) (Merck, Germany). The resulting supernatants of overnight culture of representative isolates were neutralized to pH = 6.5 - 7.0, centrifuged at 14000 rpm for five min, and filtered through a 0.20 μm pore membrane (Millipore, USA). Next, wells of 6 mm in diameter were created in these agar plates and filled with 50 μL of each isolates supernatant (11). Sterile discs (6 mm in diameter) were soaked with culture filtrate of each isolate and placed on a 100 mm plate for the disc diffusion assay (12).
3.4. Acid Tolerance
In this study, representative isolates from each profile were used for evaluation of probiotic properties. Isolates were grown for six hours in MRS broth at 37°C. An aliquot of 1 mL of the six-hour old culture was inoculated in 100 mL of MRS broth, which had its pH adjusted to 3 or 7. Bacterial growth was monitored by determination of optical density at 620 nm after 24 hours of incubation at 37°C. The percentage difference between the variation of optical density at pH = 7.0 (∆OD pH = 7) and the variation of optical density at pH = 3 (∆OD pH = 3) would give an index of surviving isolates that can be expressed as follows (Equation 1):

Classification criteria of probiotics included: good if the isolate survived at pH 3 after 24 hour; poor if the isolate did not survive in any experimental condition. An isolate survived if it demonstrated a surviving percentage equal to or greater than 50% (13, 14).
3.5. Bile Salt Tolerance
The tolerance of isolates to Bile Salts (BS) was evaluated in MRS supplemented with Bile Salts. Isolate cultures were grown for six hours in MRS broth at 37°C. An aliquot of 1 mL of the six-hour old culture was inoculated in 100 mL of MRS broth with 0.4% (w/v) bile salts (Sigma, Germany). Bacterial growth was monitored by determination of optical density at 620 nm after six and 24 hours of incubation at 37°C. The percentage difference between the variation of optical density of the culture without bile salts (∆OD 0%BS) and the variation of optical density of culture containing 0.4% bile salts (∆OD 0.4%BS) would give an index of isolates surviving that can be expressed as follows (Equation 2):

Classification criteria of probiotics included: excellent if the isolate survived at 0.4% bile salt after 24 hours; very good if the isolate survived in 0.4% bile salt after six hours; poor if the isolate did not survive in any experimental condition. An isolate survived if it demonstrated a surviving percentage equal to or greater than 50% (5, 13, 14).
4. Results
4.1. Isolation and Identification of Lactic Acid Bacteria
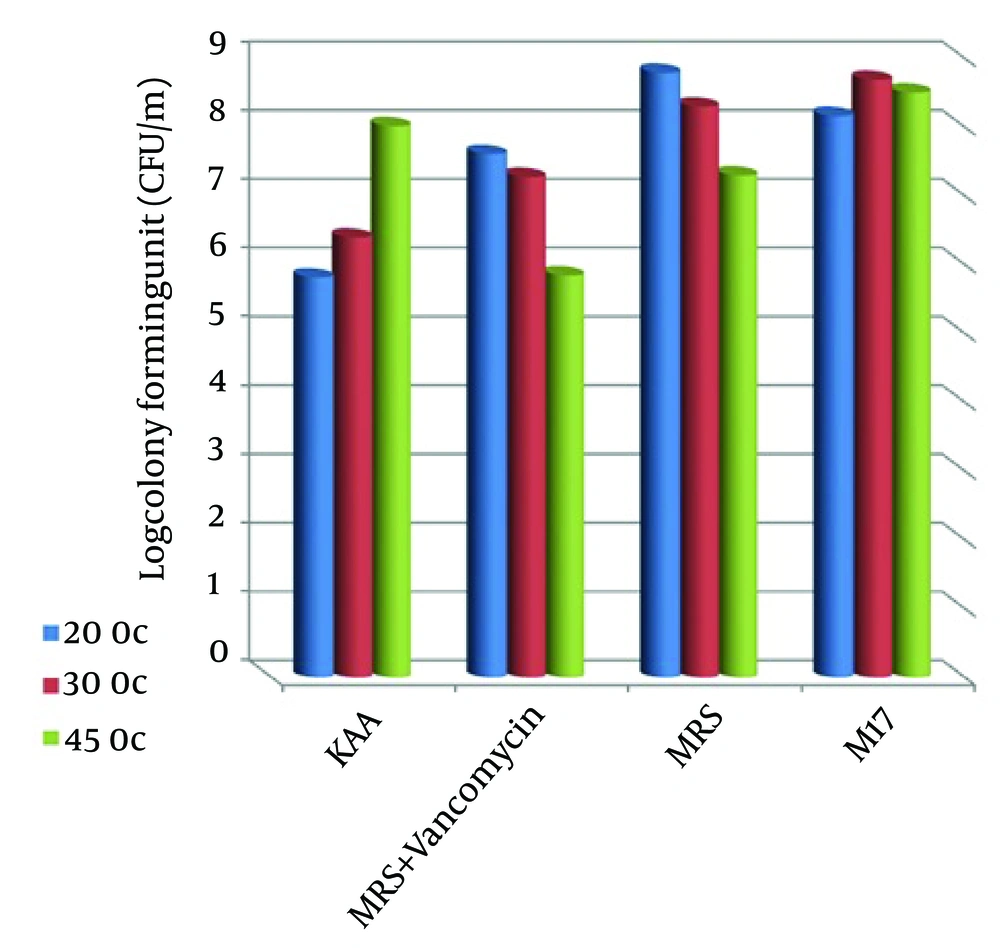
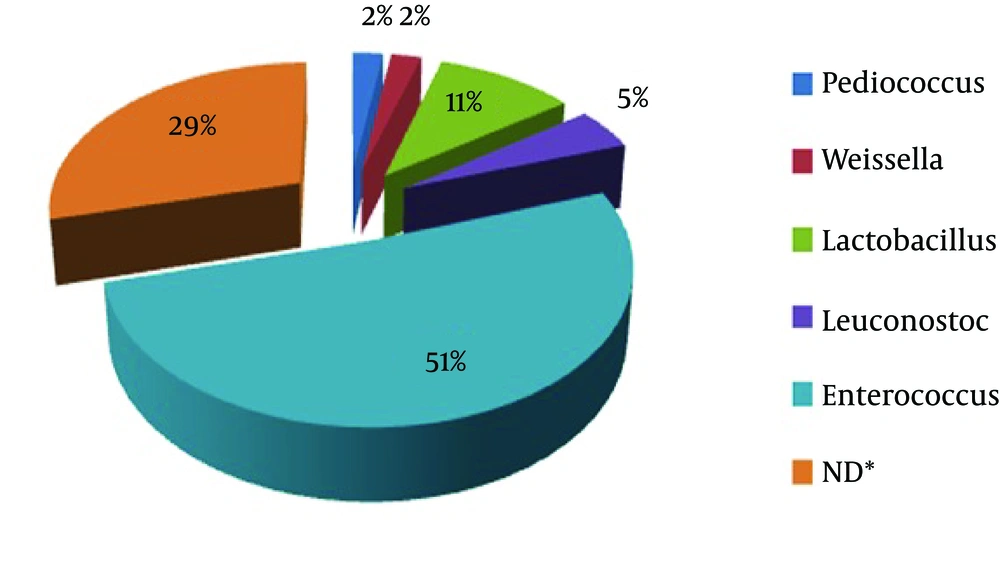
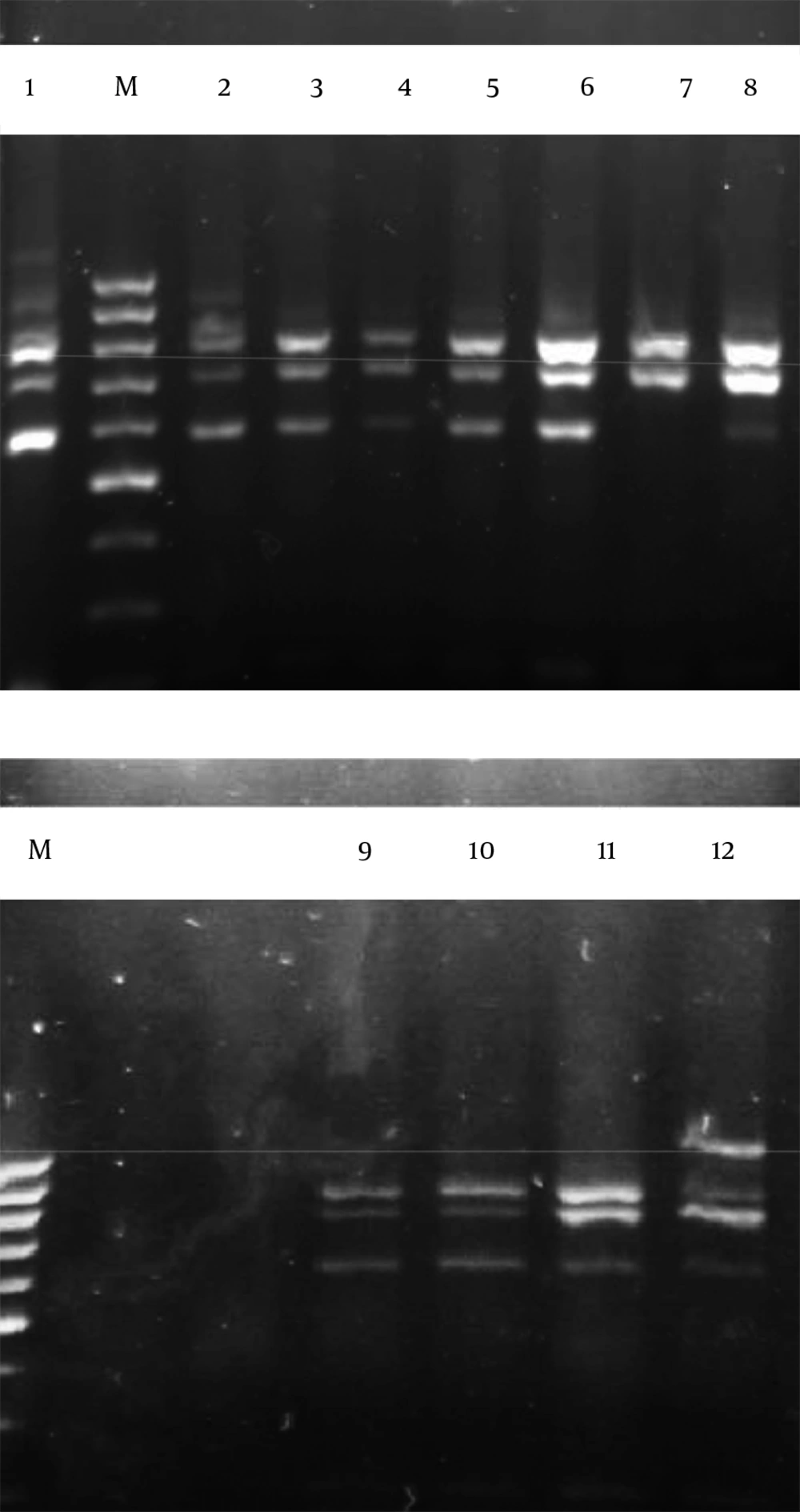
A total of 64 bacteria were isolated from camel milk samples. The results of colony counts from different media and temperatures are shown in Table 1 and Figure 1. This Figure shows that the most colony-forming units per 1 mL (8.79 ± 0.2) was obtained from MRS agar media at 20°C. All these isolates were gram positive, catalase negative rods or cocci. These strains were classified at the genus level based on biochemical tests and morphological properties. Results indicated that the 64 isolates were separated to five groups. These five groups included Leuconostoc, Lactobacillus, Enterococcus, Weissella and Pediococcus genus (Figure 2). Figure 2 showed that Enterococcus spp were dominant (51%) in comparison with other LAB genus. The restriction enzyme Taq I for restriction digestions of the amplicons were able to differentiate all 62 LAB isolates into nine restriction patterns. These ARDRA profiles are shown in Figure 3 and listed in Table 2.
| Isolation Medium | Isolation Temperature, °C | ||
|---|---|---|---|
| 20 | 37 | 45 | |
| KAA | 5.81 ± 0.02 | 6.39 ± 0.05 | 8.01 ± 0.05 |
| MRS + Vancomycin | 7.61 ± 0.07 | 7.27 ± 0.06 | 5.84 ± 0.1 |
| MRS | 8.79 ± 0.2 | 8.30 ± 0.03 | 7.30 ± 0.15 |
| M17 | 8.16 ± 0.03 | 8.69 ± 0.1 | 8.50 ± 0.04 |
a Data are presented as mean ± SD.
M; 100 bp marker, 1; E. durans strain R02-23, 2; E. faecium strain E6, 3; E. lactis, 4; E. faecium strain Y-2, 5; E. lactis, 6; E. lactis, 7; E. faecium strain JZ1-1, 8; W. cibaria, 9; P. pentosaceus strain KDLLL3-3, 10; P. pentosaceus strain KDLLL3-3, 11; L. casei and 12; L. mesenteroides strain 54.
| ARDRA, Pattern | Closest Relative | Representative Isolate | Identity a |
|---|---|---|---|
| 1 | Enterococcus durans strain R02-23 | 4-5-6-7-11-12-15-19-23-24-41-56-64 | 99 |
| 2 | Enterococcus faecium strain E6 | 8-9-14-16-21-26-31-37-40-43-55-58 | 99 |
| 3 | Enterococcus lactis | 17-18-20-30-38-51-59-60 | 98 |
| 4 | Enterococcus faecium strain Y-2 | 1-2-10-13-27-28-57-62-63 | 99 |
| 5 | Enterococcus faecium strain JZ1-1 | 22-33-50-61 | 99 |
| 6 | Weissella cibaria | 32-36 | 98 |
| 7 | Pediococcus pentosaceus | 3-52 | 99 |
| 8 | Lactobacillus casei | 25-34-35-39-42-46-47-48-49-53 | 98 |
| 9 | Leuconostoc mesenteroides | 29-44-45-54 | 99 |
a Data are presented as (%).
Representative isolates from each profile were analyzed for their 16S rDNA sequences. Homology searches of the sequences revealed (with 98 - 99% homology) that profile 1 belonged to E. durans strain R02-23, profile 2 belonged to E. faecium strain E6, profile 3 belonged to E. lactis, profile 4 belonged to E. faecium strain Y-2, profile 5 belonged to E. lactis, profile 6 belonged to E. lactis, profile 7 belonged to E. faeciumstrain JZ1-1, profile 8 belonged to L. mesenteroides strain 54, profile 9 belonged to P. pentosaceus, profile 10 belonged to P. pentosaceus, profile 11 belonged to L. casei and profile 12 belonged to W. cibaria (Figure 3). Results showed that the restriction enzyme Hha I could not differentiate among isolates based on restriction enzyme digestion profiles.
4.2. Lactic Acid Bacteria Antimicrobial Activity
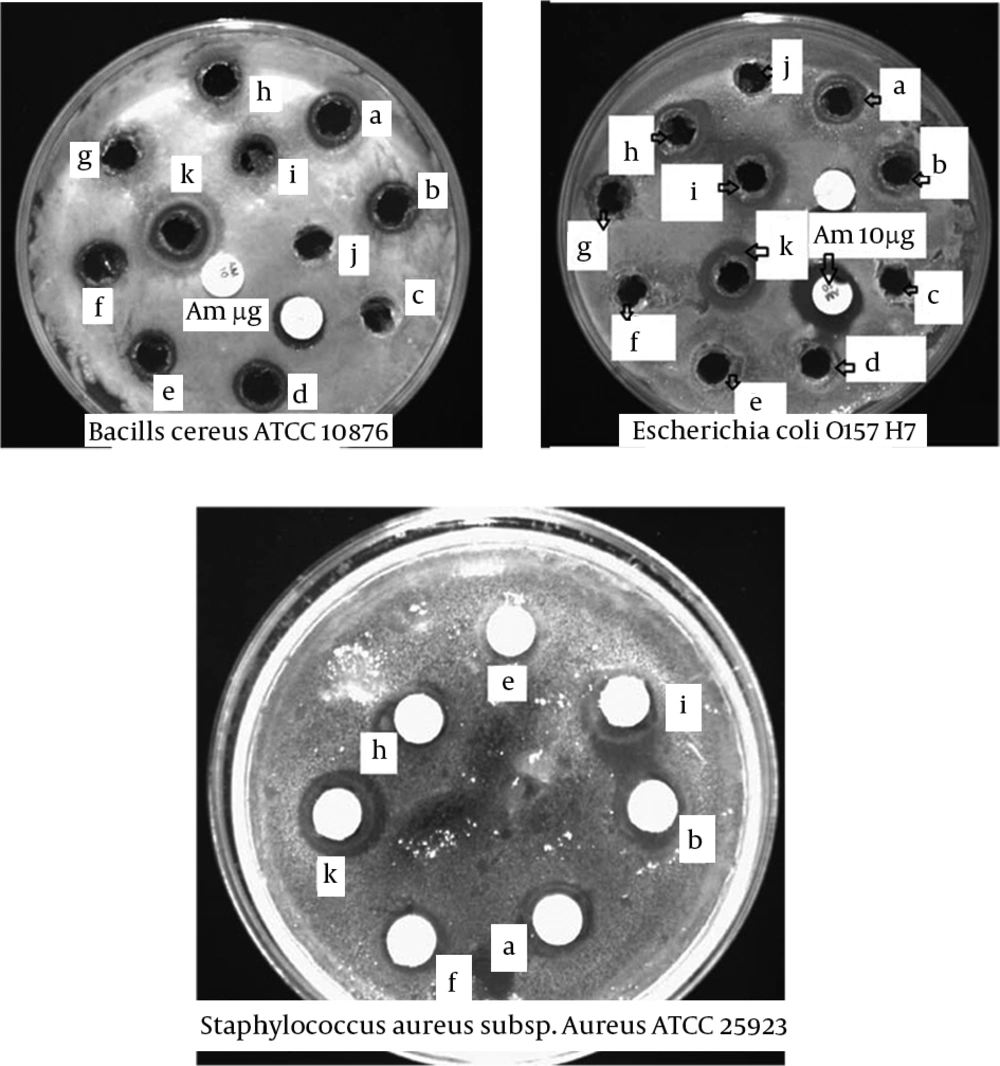
Figure 4 illustrates the zones of inhibition against pathogenic bacteria under study. In general, the studied isolates had inhibitory activity against pathogenic bacteria, because the inhibition was scored positive if the diameter of the clear zone around the colonies was 0.5 mm or larger. According to Figure 4, isolates P. pentosaceus, W. cibaria, E. faecium strain E6, E. lactis, E. durans strain R02-2, L. casei and L. mesenteroides strain 54, respectively had the highest inhibitory activity against S. aureus subsp. aureus ATCC 25923 and isolates P. pentosaceus, E. durans strain R02-2, E. faecium strain E6, E. faecium strain Y-2, L. mesenteroides strain 54, L. casei and E. lactis, respectively had the highest inhibitory spectrum against B. cereus ATCC 10876. Also, isolates P. pentosaceus, W. cibaria, E. lactis, E. durans strain R02-2, E. faecium strain E6 and E. faecium strain JZ1-1, respectively had the highest inhibitory spectrum against E. coli O157 H7. Isolate P. pentosaceus indicated the highest inhibition effect to growth indicator bacteria.
B. cereus ATCC 10876, E. coli O157 H7 (well diffusion assay) and S. aureus subsp. aureus (disc diffusion assay); a, E. durans strain R02-2; b, E. faecium strain E6; c, control - (MRS without bacteria); d, E. faecium strain Y-2; e, L. mesenteroides strain 54; f, L. casei; g, E. faecium strain JZ1-1; h, E. lactis; I, W. Cibaria; j, control – (MRS without bacteria); k, P. pentosaceus.
4.3. Acid Tolerance and Bile Salt Tolerance
Among the seven selected representative isolates under the acidic conditions using the rapid selective method, five isolates, namely E. durans, L. casei, E. lactis, P. pentosaceus and W. cibaria, showed good tolerance. Also, four isolates, namely E. durans, E. faecium, L. casei, P. pentosaceus, demonstrated excellent capacity and one isolate, namely E. lactis, demonstrated good capacity to resist bile salts by presenting a surviving percentage greater than 50% under exposure to 0.4% bile salts after six hours at 37°C (Table 3).
| Strain | 0.4% Bile Salts | pH = 3 | ||
|---|---|---|---|---|
| 6 h | 24 h | 6 h | 24 h | |
| E. duransstrain R02-23 | 51.98 | 51.75 | 88.81 | 81.10 |
| E. faecium strain E6 | 70.98 | 69.36 | 47.51 | 44.23 |
| E. lactis | 60.05 | 47.73 | 62.06 | 53.04 |
| L. casei | 58.76 | 55.81 | 78 | 75.80 |
| L. mesenteroides | 39 | 35.54 | 46.51 | 46.19 |
| P. pentosaceus | 95 | 92.34 | 98.83 | 90.96 |
| W. cibaria | 46.57 | 44.92 | 50.84 | 50.24 |
5. Discussion
This study showed that in preliminary screening of camel milk LAB community at the genus level, Enterococcus spp were dominant in comparison with other LAB genus. Because of high salt presence in camel milk compared to other livestock animals, large numbers of Enterococcus spp. can live in camel milk. It was found that the selected primers were able to amplify the ITS region between the 16S and 23S rRNA genes of each LAB isolate from camel milk. In this study, ARDRA facilitated the screening and identification of a large number 62 of LAB isolates obtained from camel milk. Also, other researchers have reported the merits of ARDRA for rapid grouping and identification of isolates of LAB (15). This study revealed that most of the isolates can inhibit the growth of S. aureus, B. cereus and E. coli, because the clear zone around the colonies was 0.5 mm or larger. Consuming camel milk and its fermented products can help human health and protect the body against occurrences of food poisoning (16).
In the point of view of probiotic potential, E. durans, L. casei, E. lactis and P. pentosaceus isolated from camel milk were selected as probiotic bacteria. In another study on camel milk, Yateem et al. (17) showed the presence of L. plantarum, L. pentosus and L. lactis as probiotic LAB in raw camel milk. This study suggests that camel milk is a potential source for the isolation of probiotic LAB strains and can be considered good for health with antibacterial properties against pathogenic bacteria because of the presence of bacteriocin-producing strains such as Enterococcus spp. Weissella spp. and Pediococcus spp.