1. Background
Aromatic compounds are known as a group of highly persistent environmental pollutants (1), most of which possess toxic, carcinogenic and mutagenic properties (2). These chemicals are also recalcitrant to degradation. For example, the half-life of the hydrolyzed form of the dye compound, reactive blue 19, is about 46 years at pH 7 and 25°C (3).Para-amino acetanilide (PAA) is an aromatic compound used in the color industry; it is used as an intermediate in the manufacture of azo dyes and pharmaceuticals (4). Several physicochemical and biological methods are available for the removal of aromatic pollutants from the environment. Physicochemical approaches such as filtration, coagulation, activated carbon, photo-degradation, and chemical flocculation are used for eliminating aromatic compounds from the environment. However, physicochemical methods of removing aromatic compounds from the environment have some disadvantages such as high costs, being coupled with sludge formation, and release of toxic materials (5). Furthermore, the traditional techniques applied in textile wastewater treatment, such as chemical coagulation/flocculation, membrane separation or activated carbon adsorption are known to be costly and only transfer the pollutants from the liquid to the solid phase (6). Therefore, bioremediation is a cost-effective and environmental-friendly alternative for physical and chemical methods of cleaning environmental pollutants. Bioremediation (which is largely dependent on biotic and abiotic factors such as bacterial population size, pH, temperature, etc.) (7, 8) has been intensively studied over the past two decades, driven by the need for a low-cost, sustainable with the natural environment (9).
Among all organisms, microorganisms, especially bacteria, have been widely studied and frequently used for biodegradation strategies; since they are able to use chemical pollutants as the sole source of carbon and nitrogen (1). Some research groups have reported the biodegradation of aromatic compounds by bacteria belonging to Halomonas species (10, 11). The genus Halomonas presents a group of Gram-negative, rod-shaped and aerobic halotolerant bacteria, which are able to grow in a wide range of salt concentrations (12). Bacteria belonging to genus Halomonas have applicable potentials in various fields of industry, ecology and biotechnology (13, 14). Over the past decade, interest in Halomonas species has also focused on their ability to degrade aromatic compounds such as benzoic, salicylic, p-hydroxybenzoic, p-coumaric, cinnamic, phenylacetic and ferulic acid (15). Oie et al. (11) showed that the haloalkaliphilic bacterium, Halomonascampisalis, is able to degrade model aromatic compounds; benzoate and salicylate, at pH 9 and in up to 100 g/L NaCl. They detected metabolites, such as catechol and cis-muconate.
Located northwest of Iran, in the Azarbayjan region, Urmia Lake is the largest saline lake in the Middle East and the second largest salt water lake on the earth. Urmia Lake, like the dead-sea, is well known for its extreme water salinity. Traditionally, the lake water is believed to possess healing properties such as curing rheumatism. The NaCl concentration of Urmia Lake was determined to be about 34 g/L in 1915, which has increased to more than 300 g/L because of drought, evaporation, and higher demands for agricultural water in the lake’s basin. This continuing development has simultaneously become a major concern and the survival of the lake has been a critical and challenging issue in the recent years. Such an evolution encouraged us to study the halophilic bacterial population of Urmia Lake resulting in isolation of some new Halomonas isolates including Halomonas sp. TBZ3 (16).
2. Objectives
Considering the environmental problems posed by aromatic compounds such as PAA (used by various factories in the area) and the ability of Halomonas bacteria to degrade aromatic compounds (10, 11), this study aimed to characterize the TBZ3 isolate, and to elucidate its ability as a biodegradative agent to decompose PAA. Additionally, some factors affecting biodegradation rate (temperature, pH and salinity) were optimized using the response surface methodology (RSM) method.
3. Materials and Methods
3.1. Description of Halomonas sp. TBZ3
Halomonas sp. TBZ3 was isolated from water samples of the Urmia Lake. Based on 16S rRNA gene sequence (EU305729 accession number), TBZ3 belongs to the Halomonas genus. These bacteria are Gram-negative, oxidase- and catalase-positive. Its colonies are convex, smooth and translucent creamy. Growth occurs in a medium with 7.5% - 20% (w/v) NaCl. No growth occurs in the absence of salt. It grows at temperatures between 10 and 45°C and in pH ranging from 6 to 9. It does not produce acid from D-glucose, D-mannose, D-fructose, maltose, D-mannitol, rhamnose, sucrose, D-trehalose, D-galactose, cellobiose, D-raffinose, xylose, lactose, ribose, arabinose, D-salicin and aesculin. Indole and urease activities, and H2S production are negative. Phenylalanine-deaminase production is positive. Tween 20 and Tween 80 are hydrolyzed, yet tyrosine, starch and casein are not hydrolyzed. Nitrate is not reduced to nitrite. Arginine is not deaminated. In order to further characterize and confirm that the TBZ3 isolate is not identical with other known bacterial type strains; the isolate was subjected to complementary characteristic studies (16).
3.2. DNA-DNA Hybridization and DNA G + C Content
Primarily, DNA-DNA Hybridization (DDH) between TBZ3 and reference isolate Halomonasdenitrificans DSM18045T, was carried out as described by De Ley et al. (17) with the modifications described by Huss et al. (18), by German collection of microorganisms and cell cultures (DSMZ). The DNA-DNA hybridization between TBZ3 and Halomonas saccharevitans LMG 23976T was done by a Belgian coordinated collection of microorganisms, laboratory of Microbiology of Gent (BCCM/LMG), Gent, Belgium. Hybridization was performed in the presence of 50% formamide at 50°C, according to a modified version of the method described by Ezaki et al. (19).
3.3. Culture Media and Aromatic Compounds
Para-Amino Acetanilide was provided by the Boyakh Saz Company. At first, the previously isolated TBZ3 cells were cultured in marine Broth 2216 (Difco) and incubated at 30°C to reach a suitable state of growth. Then, 400 μL aliquots of fresh culture (OD 0.5) were inoculated in Mineral Medium (MM) pH 7.0 to conduct the biodegradation process. This medium contained (per liter): K2HPO4 (8.0 g), KCl (1.0 g), NH4Cl (1.5 g), MgSO4 (0.2 g) and NaCl (70.0 g), and was supplemented with 40 ppm of filter sterilized (0.45 μm) PAA as the sole source of carbon (20, 21). Biodegradation tests were conducted in series consisting of, I) MM + PAA + bacterial isolate; II) MM + PAA, and III) medium + bacterial isolate; with (II) and (III) samples serving as controls (22).
3.4. Para-Amino Acetanilide Biodegradation Capacity of Halomonas sp. TBZ3
To evaluate the PAA degradation ability of Halomonas sp. TBZ3, this substrate was provided with a concentration of 40 ppm as the sole source of carbon. Then, the samples were incubated in a 100 rpm Vision (VS-8480) shaking incubator at 30°C. Screening was done through spectroscopic scanning of the samples using a spectrophotometer (UNICO UV-1200) at wavelengths of 200 - 800 nm with λmax of 258 nm. Before starting this spectroscopic operation, samples were centrifuged at 13000 rpm for 15 minutes. The spectrophotograms of the samples before and after the incubation were compared. All experiments were done at least in triplicates (23).
3.5. Quantitative Estimation of the Process
Quantitative estimation was accomplished for PAA bacterial biodegradation. Para-Amino Acetanilide was fed to the bacterial isolate, at a concentration of 40 ppm, as the sole source of carbon. The absorbance was measured at 258 nm using the UNICO VU-1200 spectrophotometer. The absorbance was used as a measure of biodegradation rate. The biodegradation rate was estimated using Equation 1.
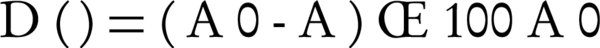
Where, A0 is the absorbance value of the control sample, A is the absorbance value after degradation, and D (%) is the biodegradation percentage (23).
3.6. Biodegradation Metabolites Detection
Detection of metabolites produced during PAA biodegradation was carried out using gas chromatography-mass spectroscopy (GC-MS). Analysis of GC-MS was conducted by GC-MS Agilent 6890 (USA) equipped with a 30 m × 0.25 mm × 25 μm HP-5MS capillary column coupled with an Agilent 5973 mass spectrometer (Agilent Technologies, Palo Alto, CA) operating in EI mode at 70 eV with the following features: helium as the carrier gas with a pressure of 34 psi at injection port and a quadrupole filter. The injection port temperature was 280°C. The initial oven temperature was 50°C and post-run temperature was 300°C (21, 24).
3.7. Optimization of Biodegradation Conditions
The RSM method was exploited to optimize the biodegradation conditions and for this purpose, Minitab 15 software was employed. Three independent variables (pH, temperature and concentration of salt) were selected as they were expected to affect biodegradation efficiency. Eight (23) factorial points and six (2 × 3) axial points with six replicates at the center point, thus a total of 20 experiments, were conducted in a randomized order. All of the experiments were done at least in triplicate. The center point replicates were selected to verify any change in the assessment procedure, as a measure of exactitude property. A full quadratic model for this design is given as Equation 2:
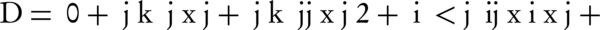
Where, D is the degradation efficiency, xi and xj are variables (i and j = 1 to k), β0, βj, βjj and βij are the constant term, coefficients of linear, quadratic and second-order terms, respectively, ε is the error, and k is the number of independent variables (here = 3). Each variable was coded at five levels between -2 and + 2, according to the Equation 3 (25).
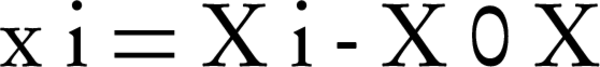
Where, xi is the coded value of the variable, Xi is the existent value of the variable, X0 is the center point value, and δX is the step change between the levels. Following initial investigations and determination of efficient parameters in the biodegradation procedure, the model was expressed as Equation 4 (25).
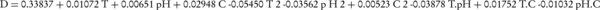
At confidence level of 95%, only the linear terms, T2 and T. pH had a meaningful effect on the results. Therefore, considering this point, the optimum conditions (for the maximum biodegradation efficiency) were predicted by the software. Five levels of temperature, pH and salinity were selected to reach the optimum values for the biodegradation process. These values for each variable were selected based on primary results gained from preliminary experiments and the reports of related scientific literature. Table 1 demonstrates the experimental design procedure.
| Standard Order | Temperature, °C | pHZ | C, % w/v |
|---|---|---|---|
| 1 | 25 (- 1) | 6.5 (- 1) | 3.5 (- 1) |
| 2 | 35 (1) | 6.5 (- 1) | 3.5 (- 1) |
| 3 | 25 (- 1) | 7.5 (1) | 3.5 (- 1) |
| 4 | 35 (1) | 7.5 (1) | 3.5 (- 1) |
| 5 | 25 (- 1) | 6.5 (- 1) | 10.5 (1) |
| 6 | 35 (1) | 6.5 (- 1) | 10.5 (1) |
| 7 | 25 (- 1) | 7.5 (1) | 10.5 (1) |
| 8 | 35 (1) | 7.5 (1) | 10.5 (1) |
| 9 | 20 (- 2) | 7.0 (0) | 7.0 (0) |
| 10 | 40 (2) | 7.0 (0) | 7.0 (0) |
| 11 | 30 (0) | 6.0 (- 2) | 7.0 (0) |
| 12 | 30 (0) | 8.0 (2) | 7.0 (0) |
| 13 | 30 (0) | 7.0 (0) | 0.0 (- 2) |
| 14 | 30 (0) | 7.0 (0) | 14.0 (2) |
| 15 - 20 | 30 (0) | 7.0 (0) | 7.0 (0) |
a The values between parenthesis are related to coded values.
Maximum absorbance at 258 nm was evaluated under these conditions using a spectrophotometer for measuring biodegradation rate. Response Surface Methodology is used to optimize more than one parameter simultaneously; parameters related to the biodegradation process are optimized one by one. For instance, Dehghani et al. (26) optimized pH in alachlor biodegradation. Response Surface Methodology naturally includes a proper number of repetitions, yet to ensure accuracy, all of the optimization experiments were done in triplicates (10, 27).
4. Results
4.1. Halomonas sp. TBZ3 DNA G + C Content and DNA-DNA Relatedness
The G + C content of the genomic DNA of Halomonas sp. TBZ3 was 67.1 mol%. The DNA-DNA hybridization experiments between Halomonas sp. TBZ3, H. denitrificans DSM 18045T and H. saccharevitans LMG 23976T revealed relatedness levels of 57% and 65%, respectively.
4.2. Quantitative Examination of Biodegradation
Aromatic compounds are stable chemicals and are slowly degraded in the environment. However, biodegradation of these compounds using Halomonas species has already been shown (10, 15). Accordingly, primary investigation of PAA biodegradation by the TBZ3 isolate was conducted. To achieve this, PAA was added to the culture medium as the sole carbon source and its biodegradation was measured using spectrophotometric techniques. Spectrophotometric analysis showed that isolate TBZ3 is able to degrade PAA. Comparative results indicated that TBZ3-mediated biodegradation rate of PAA was 32.91% after a one-week incubation period.
4.3. Biodegradation Metabolites Detection
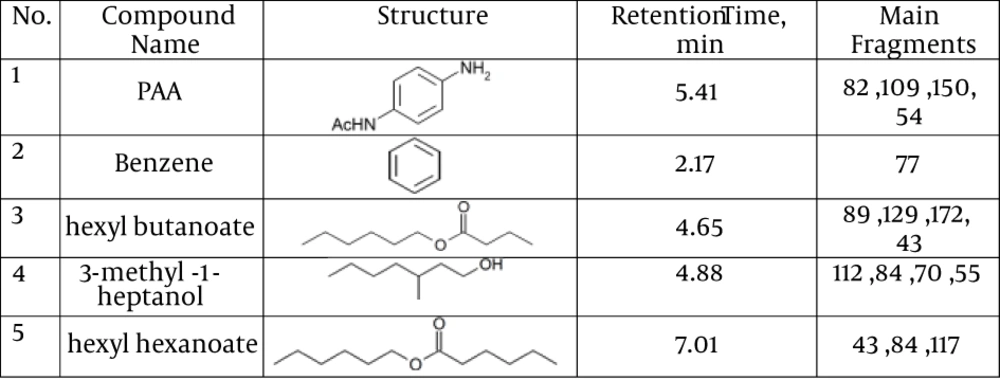
Detection of PAA biodegradation products was done by the GC-MS method. The GC-MS results showed that PAA is degraded by TBZ3. Furthermore, GC-MS revealed products at retention times of 2.17, 4.65, 4.88, 5.41 and 7.01 minutes. Mass spectroscopy data demonstrated that the biodegradation metabolites of PAA by TBZ3 were benzene, hexyl butanoate, 3-methyl-1-heptanol and hexyl hexanoate. The results of GC-MS analysis are summarized in Figure 1.
4.4. Optimization of Biodegradation Conditions
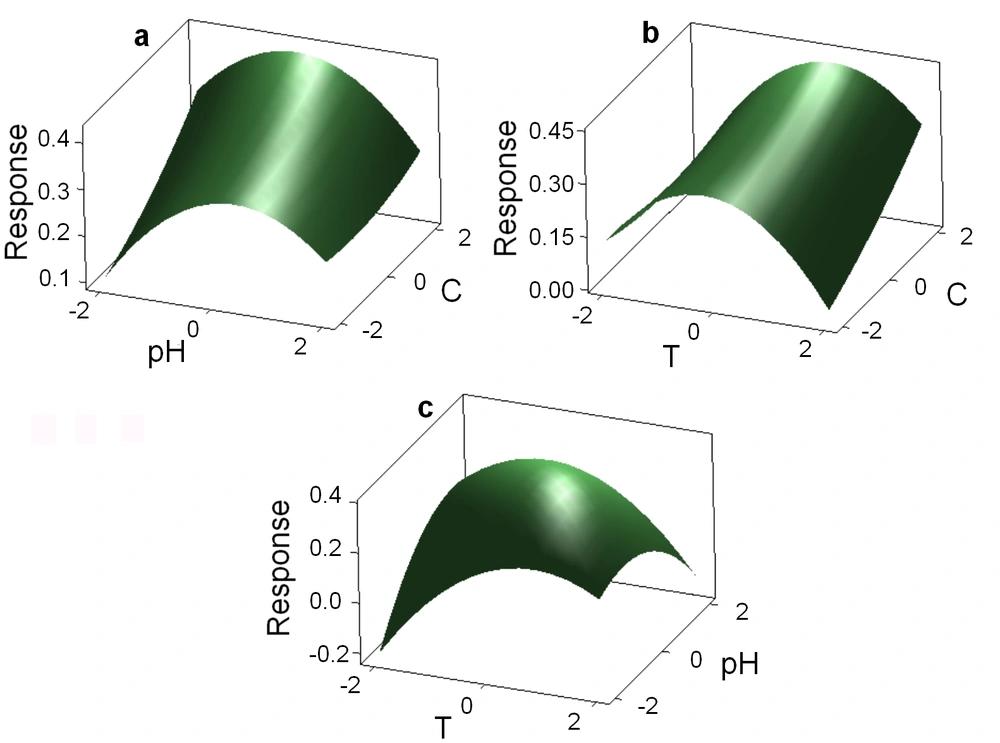
Response Surface Methodology was applied to find the optimum conditions for biodegradation temperature, pH, and salinity percentage. The spectrophotometric curves of TBZ3-treated PAA samples and controls were compared. The response surface plots are shown in Figure 2. The results showed that PAA is biodegraded by isolate TBZ3. It was found that temperature of 32.92°C, pH 6.76 and salinity of 14% were the optimum conditions for PAA biodegradation.
5. Discussion
Bioremediation provides a low-cost and environmentally safe way to remove pollutants from the environment (8). Aromatic compounds are stable chemicals and are slowly degraded in the environment. Biodegradation of these compounds using Halomonas species has already been shown (11, 15). In this study, a native, newly isolated Halomonas strain was exploited for biodegradation of an aromatic compound used in the area. Similar studies have used native bacteria for bioremediation goals. For example, Moghadam et al. (8) used bacteria isolated from Nayband Bay region for biodegradation of phenanthrene. In another study, Abari et al. (7) introduced a novel bacterial isolate capable of toluene biodegradation. Taking into account that relatedness levels significantly below the threshold value of 70% are generally accepted for species delineation (28, 29), the DDH values of TBZ3 justify distinction of strain TBZ3 from H. denitrificans DSM18045T and H. saccharevitans LMG 23976T at the species level.
Mass spectroscopy data helped us identify the biodegradation metabolites of PAA by TBZ3. In some studies, GC-MS has been applied to determine metabolites produced during biodegradation of aromatic compounds. For example, Lin et al. (21) identified degradation metabolites of naphthalene by isolate Bacillus fusiformis using GC-MS. Khataee et al. (29) also used the GC-MS method for detection of metabolites produced during the degradation of basic red 46. Abari et al. (7) used gas chromatography as a sensitive and accurate method to follow up toluene biodegradation rate by bacteria isolated from wastewater, and another study by Moghadam et al. used the same method to show that phenanthrene can be degraded by bacteria isolated from coastal sediments of Nayband bay in Iran (8). Apart from revealing aromatic compound degradation capability, biodegradation metabolites were also detected in this study (such as benzene, hexyl butanoate, 3-methyl-1-heptanol and hexyl hexanoate).
According to RSM results, temperature of 32.92°C, pH 6.76, and salinity of 14% were the optimum conditions for biodegradation of PAA by Halomonas sp. TBZ3 with a confidence level of 95% (at level α = 0.05). The experiments showed that biodegradation of PAA is efficiently increased under these optimum conditions. After optimization, biodegradation rate rises from 32.91% to about 66%. In the conventional optimization methods, the conditions are optimized by changing only one parameter and keeping the others unchanged (“one at a time” method). Apart from being time consuming, this method may not lead in accurate results. In most studies, only one or two parameters involved in biodegradation are examined. For instance, Dehghani et al. (26) optimized some parameters influencing alachlor biodegradation such as pH. They examined these factors individually, changing one factor and keeping the others unchanged (27).
Response Surface Methodology naturally includes a proper number of repetitions, yet the data analysis showed that the additional repetitions conducted in this study could lead to further accuracy. Therefore, it seems that the use of RSM and examination of more parameters is a better solution for the optimization of biodegradation conditions (29). Salinity of 14% determined at the optimization stage was in accordance with the salinity found for the optimum growth conditions of Halomonas sp. TBZ3 (7.5% - 20%). The optimum pH of 6.74 found for biodegradation of para-amino acetanilide was also in accordance with the optimum growth pH of the Halomonas sp. TBZ3 (pH 6 - 9) (16).
In conclusion, our findings showed that Halomonas sp. TBZ3 isolated from the Urmia Lake of Iran is able to degrade PAA. Optimum biodegradation conditions were determined through RSM as an efficient and accurate method. These optimum conditions were in accordance with optimum growth conditions and increased biodegradation rate remarkably. Further studies regarding potential of Halomonas sp. TBZ3 in biodegradation of other aromatic compounds are suggested.