1. Background
Antibiotic resistance emergence in pathogenic bacteria in each of hospital and community as well, represents a significant public health problem (1-4). Inheritance of this phenomenon in bacteria is controlled by either chromosome or plasmid according to many studies. Bacterial cells are able to transfer genes horizontally, which can occur in three ways; through plasmids, phages, or uptake of naked DNA “transformation” (5). Plasmids as extra-chromosomal pieces of DNA are capable to replicate independently of the genome. Plasmids have a considerable role in resistance to many antibiotics and in spreading of antibiotic resistance genes (6-12). These cause serious problems, since plasmids can cross many species and genus barriers, which allow resistance to spread and persist in organisms not necessarily subject to antibiotics (13).
The antibiotics resistant bacteria were isolated, not only from environments contaminated with antimicrobial agents, e.g. hospitals, fish farms, sewage effluents, and wastewater (14), but also from apparently nonselective environments (15). Kruse and Sorum (14) reported that environmental contamination with resistant bacterial pathogens is a real threat, not only as a source of disease, but also as a source from which plasmids can easily spread to other pathogens. Methicillin is a semisynthetic derivative of penicillin. It was first produced in the late 1950s and developed as a type of antibiotic called a penicillinase-resistant penicillin. It contained a modification to the original penicillin structure made it resistant to a bacterial enzyme called penicillinase (beta-lactamase). Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important hospital associated (nosocomial) bacterial pathogens worldwide. Detection of methicillin resistance among Staphylococcus isolates is based on phenotypic assays, until that confirmed genetically based on the detection of mecA gene (16), that associated with methicillin resistance S. aureus (17).
2. Objectives
The performance of mecA gene detection in non-staphylococci bacteria was examined, to clarify if it gained this gene (mecA gene) through plasmid, phage, or transformation processes, or it (mecA gene) may correlated only to S. aureus strains.
3. Materials and Methods
3.1. Sample Collection
Two sampling campaigns were conducted in October 2011 and March 2012 to collect 18, 26 and 26 water samples from river Nile, treated raw water and groundwater respectively from seven locations (Tema, Tahta, Sakolta, Sohag, Akhmim, Girga, Dar-Elsalam) along Sohag governorate. Samples were transported in icebox to the laboratory, and microbiological determinations were performed within six hours.
3.2. Bacteria
Thirty two strains were isolated from referred different water resources in Sohag governorate, Egypt and identified according to Bergey's Manual, 1989 (18). For determination of total bacterial count, serial diluted samples were grown on standard method agar (P. C. A) Acc.to APHA and ISO 4833, while Pseudomonas isolation agar medium was used for isolation of Pseudomonas aeruginosa (19). M-Endo Agar LES (Difco, USA) according to Tajima et al. (20), standard methods for examination of water and waste water were used for enumeration of total coliforms in water by membrane filter technique. Lauryl tryptose broth (Difco, USA) was used for verification of total coliforms. MFC Agar Base was used with Rosolic Acid in cultivating and enumerating fecal coliforms by the membrane filter technique. Azide dextrose broth medium was used for counting fecal streptococci (20). Aesculin Azide Agar was used for verification of fecal streptococci, and Escherichia coli was counted on MacConkey agar medium after incubation at 44°C for 48 hours. X.L.D Agar medium was used as selective medium for isolation of Salmonella spp., Shigella spp. and incubated for 24 hours at 37°C (ISO, 1978). S. aureus strain was selected and used as standard for MecA gene screening.
3.3. Antibiotics Susceptibility
All isolates of Salmonella, Shigella, Pseudomonas and E. coli were tested for susceptibility against the following antibiotics: amoxicillin/clavulanic acid (30 µg), Rifamycin (30 mcg), Cefalexin (30 mcg), Erythromycin (15 mcg), Ceftazidim (30 mcg), Norfloxacin (10 µg), Ciprofloxacin (5 µg), Cefepime (30 mcg), Ertapenem (10 µg) and Cefoxitin (30 mcg), by the Bauer’s disk diffusion method (21).
3.4. DNA Extraction
Genomic DNA was extracted according to Maloy, (22), whereas plasmids DNA were extracted according to Birnboim, (23).
3.5. Amplification for mecA Gene
The method was performed according to Towner et al. (24). DNA samples were diluted to concentration of 20 ng/µL prior to amplification. Five µL aliquot of bacterial genomic DNA was combined with 12.5 µL of master mix (500 mM KCl, 100 mM tris-HCl (pH at 25°C), 15 Mm MgCl2, 1% Triton X-100), 1 µL of a deoxynucleoside triphosphate (dNTP) mixture (concentration of dNTP, 5 mM), and 2 µL each of two 15-base oligonucleotide primers (primer MecA1 and MecA2 (concentration, 50 ng/µL)) 3.5 µL deionized water; the final volume was 25 µL.
3.6. Primers
MecA1 (5'-GTA GAA ATG ACT GAA CGT CCG ATA A)
MecA2 (5'-CCA ATT CCA CAT TGT TTC GGT CTA A)
The PCR cycling conditions were as follows: initial denaturation at 94°C for 4 minutes, followed by 30 cycles of 45 seconds at 94°C, 45 seconds at 50°C, and 60 seconds at 72°C with the final extension step at 72°C for two minutes. Ten-microliter was loaded onto agarose gel electrophoresis.
4. Results
4.1. Bacteria
As shown in Table 1, 32 bacteria were isolated from Nile and ground water. These strains were identified by means of standard procedures described in the Bergey's manual of systematic microbiology and standard laboratory procedures. Ten strains of E. coli, 10 strains of Shigella flexneri, 3 strains of S. sonnei, 2 strains of Salmonella serovar Newport, and 7 strains of P. aeruginosa were detected. No pathogenic bacteria were isolated from treated tap water.
| Isolate | Bacteria | Source | Antibiotics | Plasmid | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | CL | Amc | FEP | RF | Cip | NOR | ETP | CAZ | FOX | 1 | 2 | 3 | 4 | 5 | 6 | 7 | No. of Plasmids | |||
| AB1 | S. s. Newport | Nile-Sohag | I | R | I | R | R | S | S | R | S | R | 2000 | 1500 | 910 | 690 | R to 50% | 4 | ||
| AB2 | S. s. Newport | Nile-Akhmim | I | R | I | S | R | S | S | R | S | R | R to 40% | - | ||||||
| K1 | Sh. flexneri | Nile-Tahta | R | R | S | R | R | S | S | S | R | S | 1450 | 900 | 900 | 620 | 460 | 330 | R. to 50% | 6 |
| K2 | Sh. flexneri | Nile-Tahta | R | R | S | R | R | S | S | S | R | S | 1450 | 900 | 900 | 620 | 460 | 330 | 6 | |
| K3 | Sh. flexneri | G. w. -Sohag | R | R | I | R | R | S | R | R | R | R | 2000 | R.to 80% | 1 | |||||
| K4 | Sh. flexneri | G. w. -Sohag | R | R | S | I | R | S | S | S | R | R | 1450 | 1200 | 900 | 620 | 460 | 330 | R. to 50% | 6 |
| K5 | Sh. flexneri | Nile-Sohag | R | R | R | R | R | S | S | S | R | R | 1450 | 1200 | 900 | 620 | 460 | 330 | R. to 70% | 6 |
| K6 | Sh. flexneri | Nile-Sohag | R | R | R | R | R | S | S | S | R | S | 1450 | 1200 | 900 | 620 | 460 | 330 | R. to 60% | 6 |
| K7 | Sh .flexneri | Nile-Sohag | R | R | S | R | R | S | S | R | R | R | 1450 | 1200 | 900 | 620 | 460 | 330 | R. to 60% | 6 |
| K8 | Sh. flexneri | Nile-Sohag | R | R | R | R | R | S | S | S | S | S | R to 50% | - | ||||||
| K9 | Sh. flexneri | Nile-Girga | R | R | S | S | R | I | S | R | R | R | 1450 | 1200 | 900 | 620 | 460 | 330 | R. to 60% | 6 |
| K10 | Sh. flexneri | Nile-Sohag | R | R | S | R | R | R | R | R | R | I | 1450 | 1200 | 900 | 620 | 460 | 330 | R. to 80% | 6 |
| K11 | Sh. sonnei | Nile-Sohag | R | R | R | R | R | R | R | R | R | R | R to 100% | - | ||||||
| K12 | Sh. sonnei | Nile-Tema | R | R | S | R | R | S | R | S | R | R | 920 | 510 | R to 70% | 2 | ||||
| K13 | Sh. sonnei | Nile-Tahta | I | S | S | I | R | S | S | S | S | S | 920 | 510 | 80 | 50 | R to 10% | 4 | ||
| DK1 | E. coli | Nile-Sakolta | S | S | S | R | S | S | S | S | R | S | R. to 20% | - | ||||||
| DK2 | E. coli | Nile-Sohag | S | S | R | R | S | S | S | S | R | R | R. to 40% | - | ||||||
| DK3 | E. coli | Nile-Akmim | I | S | S | R | S | S | S | S | R | S | 1030 | R to 20% | 1 | |||||
| DK4 | E. coli | Nile-Girga | R | I | R | S | R | S | S | R | S | S | 910 | 620 | R to 40% | 2 | ||||
| DK5 | E. coli | Nile-Dar El-Salam | I | I | I | R | R | S | S | R | R | S | 1550 | 1200 | 910 | 620 | R to 40% | 4 | ||
| DK6 | E. coli | G. w. -Tema | S | S | S | R | S | S | S | S | R | S | R to 20% | - | ||||||
| DK7 | E. coli | G. w. -Tahta | S | S | S | R | S | S | S | S | R | S | R to 20% | - | ||||||
| DK8 | E. coli | Nile-Tahta | S | S | S | R | S | S | S | S | I | I | R to 10% | - | ||||||
| DK9 | E. coli | G. w. -Sakolta | S | S | S | R | S | S | S | S | R | R | R to 30% | - | ||||||
| DK10 | E. coli | Nile-Sohag | S | S | S | R | I | S | S | S | R | S | R to 20% | - | ||||||
| PSI1 | Ps. aeruginosa | Nile-Tahta | R | R | R | R | R | S | S | R | R | R | R to 80% | - | ||||||
| PSI2 | Ps. aeruginosa | Nile-Sakolta | R | R | I | S | R | S | S | S | S | S | 960 | 690 | 480 | 350 | 110 | R. to 30% | 5 | |
| PSI3 | Ps. aeruginosa | Nile-Sohag | R | R | S | R | R | S | S | S | R | R | R to 60% | - | ||||||
| PSI4 | Ps. aeruginosa | Nile-Akhmim | R | R | R | R | R | S | S | R | R | R | R to 80% | - | ||||||
| PSI5 | Ps. aeruginosa | Nile-Girag | R | R | S | R | R | S | S | S | R | R | R to 60% | - | ||||||
| PSI6 | Ps. aeruginosa | Nile-Dar El-Salam | R | R | S | R | R | S | S | S | R | R | 960 | 690 | 480 | 350 | 110 | R. to 60% | 5 | |
| PSI7 | Ps. aeruginosa | Nile-Tahta | R | R | I | R | R | S | S | R | R | R | R to 70% | - | ||||||
a Abbreviations: Amc, amoxicillin/clavulanic acid; CAZ, ceftazidime; CIP, ciprofloxacin; CL, cephalexin; E, erythromycin; ETP, ertapenem; FEP, Cefepime; FOX, cefoxitin; I, intermediate; NOR, norfloxacin; R, resistant; RF, rifamycin; S, sensitive.
4.2. Antibiotics Susceptibility
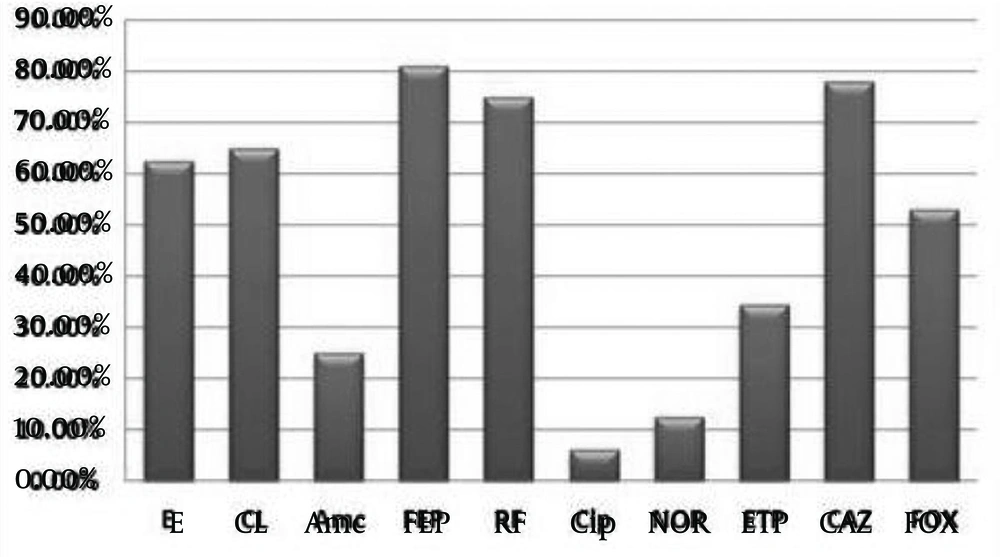
As shown in Figure 1, most of strains (81%) were resistant to Cefepime, whereas two strains only (6.25%) were resistant to Ciprofloxacin, one belonged to S. flexneri and the other to S. sonnei.
4.3. Salmonella serovar Newport
Two isolates were identified as S. serovar Newport. These strains were resistant to 40-50% of used antibiotics. The strain AB2 was resistant to 40% of antibiotics and isolated from Nile water in Akhmim region. This strain did not possess any plasmids. Strain AB1 possessed four plasmids and were resistant to Cefepime as an extra antibiotic, which inhibits the growth of the other strain, AB2 (Table 1). According to the results listed in Table 1, 50% (1 of 2) of the strains possessed plasmids (Figure 2), with resistance to about 50% of the used antibiotics, whereas the plasmids free strains were resistant to 40% of the antibiotics.
4.4. Shigella flexneri
Ten isolates were identified as S. flexneri. These strains were resistant to 50-80% of appropriate antibiotics. The strain K8, isolated from Nile water, was resistant to 50% of used antibiotics, but did not possess any plasmids. Strain K3, isolated from ground water was resistant to 80% of the antibiotics; possessed one plasmid of 2000 bp (Table 1, Figure 3). As listed in Table 1, 90% (9 of 10) of the strains possessed plasmid(s), with resistance to about 62.2% of the used antibiotics. The plasmids free strains were resistant to 50% of used antibiotics (Figure 4).
4.5. Shigella sonnei
Three isolates were identified as S. sonnei. The strain (K11) resistant to 100% of used antibiotics, was isolated from Nile water at Sohag, but did not possess any plasmids, whereas the strain K13 which isolated from Nile water at Tahta, was resistant to 10% (Cefepime) of the antibiotics, although it possessed four plasmids (Table 1, Figure 5). Strain K12 was also resistant to most of antibiotics (70%), although it had two plasmids (Table 1). In total, 66.6% (2 of 3) of S. sonnei strains included plasmids (Figure 3). These strains were resistant to 40% of the used antibiotics, whereas the plasmids free strains were resistant to 100% of the antibiotics.
4.6. Escherichia coli
Most sensitive strains were belonged to E. coli strains, since 10-40% were resistant to antibiotics. Seventy percent (70%) of the E. coli strains (n = 10) were plasmids free strains. The strain DK5 possessed four plasmids and resistant to 40% of the antibiotics, strain DK4 was resistant to 40% of used antibiotics, DK3 was resistant to 20% of antibiotics and possessed two, and one plasmids, respectively (Table 1). According to the results listed in Table 1, 30% (3 of 10) of the strains possessed plasmids, with resistance to 33.3% of the used antibiotics, whereas the plasmids free strains were resistant to 22.8% of the antibiotics.
4.7. Pseudomonas aeruginosa
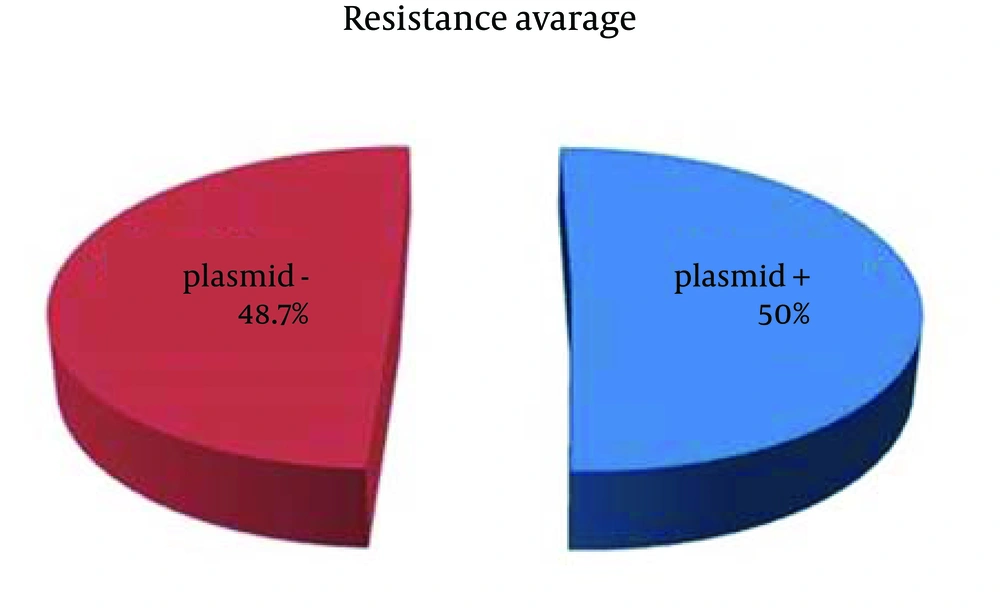
The strains resistant to 60-70% of the antibiotics (PS 11, PS13, PS 14, PS 15, PS 17), possessed no plasmids. Two strains (PS12, PS16) were identical in plasmid profile (5 plasmids), although the first strain was resistant to 30% of the antibiotics and the second strain (PS 16) to 60% of the antibiotics (Table 1). As listed in Table 1, 28.5% (2 of 7) of P. aeruginosa strains possessed plasmids (Figure 6). These plasmids including strains were resistant to 45% of the used antibiotics, whereas the others plasmids free P. aeruginosa strains were resistant to 70% of the antibiotics. According to the results listed in Table 1, 50% (16 of 32) of the total strains had plasmids. These strains showed resistance to 50% of the used antibiotics (as average value); whereas, the plasmids free strains (50%) were resistant to 48.7% of the antibiotics (Figure 5). From the results of this study, we can conclude no distinct correlation between plasmids and antibiotic resistance.
4.8. Detection of MRSA Gen by PCR Amplification
Methicillin-resistant gene was undetected (negative) in E. coli, Salmonella spp., Shigella spp. and Pseudomonas sp. as shown in Figure 7.
5. Discussion
Resistance to antibiotic is a worldwide problem. Health leaders in the world consider antibiotic-resistant bacteria as “nightmare bacteria” that “pose a catastrophic threat” to people worldwide. Among all bacterial resistance problems, Gram-negative pathogens are particularly worrisome, because they are becoming resistant to nearly all drugs considered for treatment (25). The most serious Gram-negative infections are healthcare-associated, and the most common pathogens are Enterobacteriaceae, P. aeruginosa and Acinetobacter. Treating infections of either pan-resistant or nearly pan-resistant Gram-negative microorganisms is an increasingly common challenge in many hospitals (26). The relatively high level of antibiotics resistance in pathogenic bacteria is due to the misuse of these antibiotics during treatment of bacterial infections. Hsu et al. (27) reported that differences in bacterial resistance to various antibiotics might reflect the history of antibiotic applications. Plasmid is one of the most important mediators in facilitating fast spreading of antibiotic resistance among bacteria (28).
Our results revealed that plasmid profiles differed considerably, even in the strains belonging to the same species. For example, resistance of S. serovar Newport strain AB1 to Cefepime is correlated to presence of four plasmids (Table 1, Figure 4), unlike the plasmid free strain AB2. Both of strains were resistant to the same antibiotics used in this study, although isolated from the same water source (Nile water), but different regions (Table 1). This finding assumes that there is a distinct correlation between these plasmids and resistance to antibiotics.
The strain of S. flexneri, K3 showed a unique plasmid profile (one plasmid of 2000 bp). There is no distinct correlation between plasmids and antibiotic resistance in this bacteria, since we found strains which did not possess any plasmids (strain K8), but resistant to the same antibiotics, been resistant by plasmid(s) including strains (strains K1, K2, K4) (Table 1, Figure 6). Three S. sonnei strains had different plasmid profiles. Regarding three S. sonnei strains, the more plasmid present in the bacteria, the less antibiotics resistance was found. In Table 1 and Figure 7, strain S. sonnei (K11) was resistant to 80% of the antibiotics, although it did not include plasmids, whereas the strain with four plasmids, was only resistant to 10% of used antibiotics. This can be due to this issue that these plasmids could be cryptic plasmids.
In case of plasmids including E. coli strains, the greater resistance is correlated to the presence of plasmids (40%), although, one strain (DK2) was resistant to 40%, but did not possess any plasmids (Table 1). Reinthaler et al. (29) reported that E. coli strains isolated from treated sewage were less resistant against quinolones. In P. aeruginosa, there was no distinct correlation between plasmids and antibiotic resistance, since some strains had the same plasmid profile, but showed different antibiotic sensitivity (PS12, PS16). In addition, the strains that showed resistance to a greater proportion of antibiotics did not contain plasmids (Table 1, Figure 6).
The strain origin or even the water source does not appear to have a strong effect on the plasmid profile, as shown in Table 1, since strain S. flexneri K4 isolated from ground water at Sohag region, had the same plasmid profile of other seven strains (K1, K2, K5, K6, K7, K10, K9) isolated from Nile water (Table 1). The same can be concluded from the strains of P. aeruginosa, since 5 of 7 total strains did not include plasmids, but isolated from different origins. The two strains (PS12, PS16) including the same plasmid profile, were isolated from Nile water but different origins (Sakolta, Dar El-Salam, respectively). This may be due to transfer of these strains during water movement between regions. Several studies proposed that plasmid has a positive correlation with antibiotic resistance. It is believed that the main role of plasmids that encode multiple antibiotic resistances is to confer their hosts the ability to survive in the presence of antimicrobial compounds. As an example in the pathogenic bacterium, Salmonella, plasmids of the incompatibility group HI1 accounted for a significant proportion of antibiotic resistance phenotypes.
Several studies proposed that, plasmids implicated directly in the acquisition of resistance to many antibiotics (6-12, 30), which is particularly problematic since plasmids can cross many species and genus barriers, and the rate of plasmid transfer has even been shown to increase in more heterogeneous communities (31). Plasmids allow resistance to spread and persist in niches that are not necessarily subject to antibiotics (13). In Escherichia and Shigella strains, a larger proportion of genome of plasmids codes for antibiotic resistance than that of the chromosome (32). Svara and Rankin (33) revealed that antibiotic resistance genes were significantly over-represented on plasmids, compared to on the bacterial chromosome. They also documented that environmental variation affects the evolution of plasmid-carried antibiotic resistance. Paytubi et al. (34) concluded that plasmid R27 has a strong impact on the global transcriptome of S. Typhimurium strain SL1344 when cells grow at low temperature and enter the stationary phase.
Although, multiple antibiotic resistance in bacteria is most commonly associated with presence of plasmids, which contain one or more resistance genes, each encoding a single antibiotic resistance phenotype (8, 35), some multiple antibiotic resistance are associated with the chromosome (35-38). For example, George and Levy initially described a chromosomal multiple antibiotic resistance system existed in E. coli (39). Resistance genes encoded in plasmids are often located within genetic elements called transposons. These elements include the transposase function that enables the transposon to recombine into the bacterial chromosome or plasmids (40). For this reason, the presence of plasmids in antibiotics resistant organisms is not usually recorded. The plasmid is responsible not only for antibiotic resistance, but also for other jobs, since some plasmid includes genes, which regulate pathogenicity, anaerobic respiration and metabolism determinants, heavy metal resistant. Many attempts were made to solve the antibiotics resistance problems of enteric bacteria generally. For example, Mirnejad et al. (41), identified a Lactobacilluscasei strain that strongly inhibits the development of S. sonnei and S. flexneriin vitro.
The acquisition of the mecA gene, which confers resistance to methicillin, spawning so-called methicillin-resistant S. aureus (MRSA), has resulted in a highly resilient pathogen that reached epidemic levels in many parts of the world (42). All isolated strains had broad-spectrum resistance for all tested antibiotics, especially quinolones (Ciprofloxacin) and B-Lactams (E-moxclav) as shown in Table 1. As shown in Figure 7, all strains had negative results for MRSA-PCR. Our results were in agreement with those obtained by Sekiguchi et al. (43) and Kawai et al. (44).