1. Background
Escherichia coli (E. coli), an aerobic bacterial species and the main lower intestine normal microflora, are of the leading causes of community-acquired and nosocomial infections. Virulence factors and antibiotic-resistance mechanisms are the main determinants of E. coli. These microorganisms cause urinary system infection, sepsis, peritonitis, meningitis of new-born infants, and wound infections. Use of antibiotics increased over the last decade due to non-prescription sales in many countries, frequent use in intensive care units (ICU), and use as chemotherapy in immunosuppressed and elderly patients. The increase in antibiotics use significantly increased antibiotic resistance throughout the world (1). One of the antibiotic resistance mechanisms developed by bacteria is the production of extended-spectrum-β-lactamase (ESBL), which hydrolyzes the β-lactam ring of antibiotics, including penicillins, first-, second-, and third-generation cephalosporins, and monobactams (2).
Extended spectrum β-lactamase production is phenotypically determined by the ESBL screening test, double-disk synergy test, 3-dimensional test, E-test, and automated systems (i e, Phoenix and VITEK), while genotypic determination of ESBL involves epidemiological studies, determination of the mechanism of the antibiotic resistance, and determination of clonal relationship between causative bacterial strains. In terms of epidemiological studies, it is imperative that genotypic typing of bacterial strains be implemented by rapid and highly sensitive methods in order to decrease antibiotic resistance among different strains. Phenotypically determined ESBL tests can identify isolates to be used in subsequent genotypic test, giving rise to faster results than genotypic tests (3-6).
It appears that the ESBL resistance progressed towards carbapenem resistance over time since it was first identified in 1980s in Europe (7); it nowadays necessitates more frequent genotyping of infectious strains. Therefore, pulsed-field gel electrophoresis (PFGE) is one of the most preferred genotyping methods, particularly in molecular typing of E. coli and Klebsiella species. A positive result for the presence of ESBL varies based on geographical environment as well as the method used to identify ESBL (8, 9).
2. Objectives
The current study aimed at determining both antibiotic resistance pattern and PFGE genotyping of 45 E. coli species isolated from patients hospitalized in Mevlana University Foundation Hospital (MUFH-Konya) and the Mevlana University Medical Center (MUMC) at Konya, Turkey from December 2013 to December 2014. Data presented in the current study were prospectively used to identify the most suitable antibiotic treatment for future E. coli infection cases in MUFH-Konya.
3. Methods
3.1. Ethics Statement
Ethics Committee of Mevlana University approved the study protocol (87/2015).
3.2. Patients and Clinical Specimens
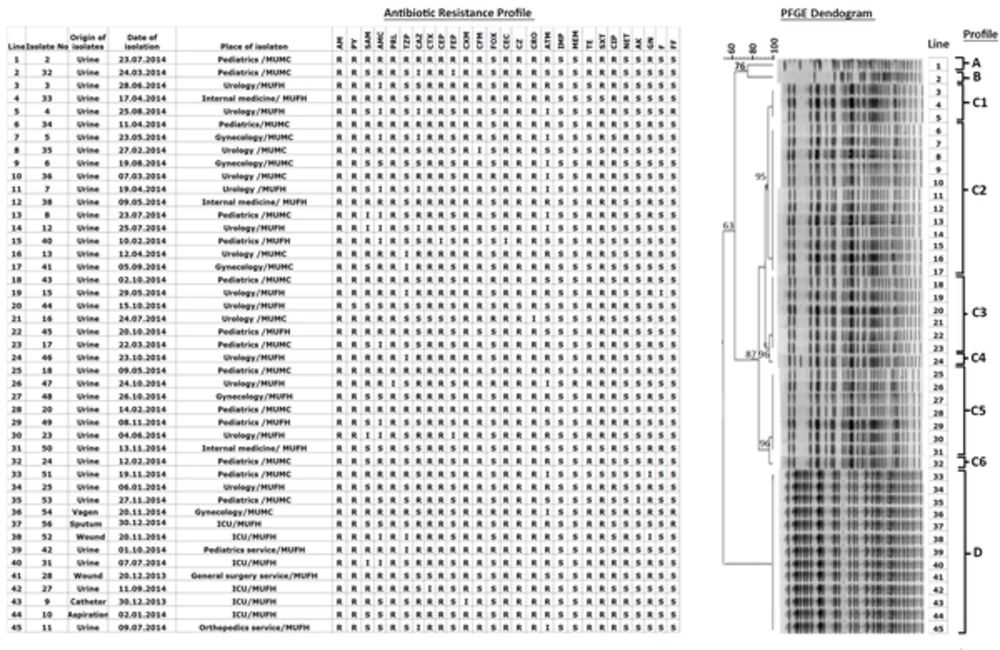
A total of 342 specimens were collected from 326 outpatients as well as 14 inpatients in a variety of units both at the Mevlana University Foundation Hospital (MUFH-Konya) and Mevlana university medical center in Konya (MUMC-Konya) from December 2013 to December 2014. ESBL-producing isolates were then stored at -20°C in the medical microbiology laboratory in MUMC-Konya. In general, 1 specimen per patient was included. Exceptionally, 4 specimens from 1 outpatient and 1 inpatient, each of 2, were also included in the current study as these specimens gave rise to different strains of ESBL-producing E. coli isolates. A total of 45 clinical isolates were obtained from 43 Turkish patients. More specifically, isolates 3 - 4 were obtained from 1 outpatient at 2-month intervals and isolates 9 - 10 were obtained from 1 inpatient at 3-day intervals (Figure 1). Extended spectrum β-lactamase producing E. coli isolates listed in lines 1 - 36 were taken from 35 outpatients, while isolates in lines 37 - 45 were taken from 8 inpatients (Figure 1). Clinical data for each patient infected with E. coli were retrieved from MUFH-Konya.
AM: Ampicillin, PY: Carbenicillin, SAM: Ampicillin-sulbactam, AMC: Amoxicillin-clavulonate, PRL: Piperacillin, TZP: Piperacillin/tazobactam, CAZ: Ceftazidime, CTX: Cefotaxime, CEP: Cefoperazone, FEP: Cefepime, CXM: Cefuroxim, CFM: Cefixime, FOX: Cefoxitin, CEC: Cefaclor, CZ: Cefazolin, CRO: Ceftriaxone, ATM: Aztreonam, IPM: Imipenem, MEM: Meropenem, TE: Tetracycline, SXT: Trimethoprim-sulfamethoxazole, CIP: Ciprofloxacin, NET: Netilmicin, AK: Amikacin, GN: Gentamycin, F: Nitrofurantoin, FF: Fosfomycin.
3.3. Isolation and Identification of E. coli Strains
Specimens were inoculated and incubated for 18 - 24 hours in the microbiology laboratory at MUMC-Konya by the conventional methods according to the clinical and laboratory standards institute (CLSI) guidelines (8). Extended spectrum β-lactamase positive E. coli isolates were identified by the initial screening test. Inhibition zones ≤ 22 mm for ceftazidime (CAZ, 30 µg), ≤ 27 mm for aztreonam (ATM, 30 µg), ≤ 27 mm for cefotaxime (CTX, 30 µg), and ≤ 25 mm for ceftriaxone (CRO, 30 µg) were considered as ESBL production. Confirmation tests were implemented by double-disk synergy and combination disk methods according to CLSI guidelines (8).
3.4. Antibiotic Resistance Studies
Antibiotic resistance profile of E. coli isolates was determined in MUMC-Konya by the Kirby-Bauer disk diffusion method according to the CLSI guidelines (8), which included a series of antibiotic disks (Bioanalyse, Ankara, Turkiye) containing amikacin (AK, 30 µg), ampicillin (AM, 10 µg), ampicillin-sulbactam (SAM, 10 µg/10 µg), amoxicillin-clavulonate (AMC, 20 µg/10 µg), aztreonam (ATM, 30 µg), ceftazidime (CAZ, 30 µg), cefotaxime (CTX, 30 µg), cefoperazone (CEP, 75 µg), carbenicillin (PY, 100 µg), cefepime (FEP, 30 µg) cefuroxime (CXM, 30 µg), ciprofloxacin (CIP, 5 µg), cefixime (CFM, 5 µg), cefoxitin (FOX, 30 µg), cefaclor (CEC, 30 µg), cefazolin (CZ, 30 µg), ceftriaxone (CRO, 30 µg), fosfomycin (FF, 50 µg), gentamycin (GN, 10 µg), imipenem (IMP, 10 µg), meropenem (MEM, 10 µg), nitrofurantoin (F, 300 µg), netilmicin (NET, 30 µg), piperacillin (PRL, 100 µg), piperacillin/tazobactam (TZP, 100 µg/10 µg), tetracyclin (TE, 30 µg), trimethoprim-sulfamethoxazole (SXT, 23.75 µg/1.25 µg), ceftazidime/clavulonic acid (CZC, 30 µg/10 µg), and cefotaxime/clavulonic acid (CTC, 30 µg/10 µg). Escherichia coli ATCC 25922 was used as the control strain in the antibiotic susceptibility tests. The isolates were classified as resistant (R), intermediate-resistant (I), or susceptible (S).
3.5. Pulsed-Field Gel Electrophoresis
Pulsed-Field Gel Electrophoresis was implemented on a BioRad CHEF-DR III PFGE System (Bio-Rad, Hercules, CA USA) at the Biotechnology Institute at Ankara University, Ankara, Turkey.
3.5.1. Preparation of Samples
Bacterial isolates were suspended in 3 mL cell suspension buffer (CSB) solution. These cells were then centrifuged at 2500 g at 4°C for 15 minutes, followed by pipetting out the CSB solution. The cell pellets were added with 1 mL of CSB solution and mixed by pipetting in and out several times. The cell concentrations were adjusted to absorbance 1 at 590 nm by UV/V in spectrometer (Beckman Coulter, USA). A 100-μL aliquot of preheated 2% low-melting temperature agarose was pipetted into 1.7 mL vials mounted on a water bath (45 - 50°C). A 100-μL aliquot of each bacterial suspension was pipetted into each Eppendorf tube and placed in a water bath. Finally, 1 mL of sodium dodecyl sulfate (SDS) solution (10%) was added to the Eppendorf tubes and mixed slowly by pipetting in and out several times.
Bacterial isolates in broth medium were carefully transferred into little molds (10 mm × 5 mm × 1.5 mm, Bio-Rad, Hercules, CA USA), which were then cooled down at 4°C for 10 minutes until homogenously solidified. The agarose molds were taken out of their molds and transferred into 5-mL tubes containing 0.5 mL cell lysis solution (CLS)-I and shaken at 37°C for 1 hour in a water bath. After removal of CLS-I from the tubes, 0.5 mL of CLS-II was added into each tube, followed by shaking at 50°C for 0.5 hour in a water bath. When CLS-II was completely removed, the agarose molds were washed 3 times with TE buffer at 50°C for 3 hours. After removal of TE buffer, 100 μL of XbaI restriction enzyme was added to the tubes with agarose molds in TE buffer and incubated at 37°C for 2 hours. After removal of the enzyme buffer, the agarose molds were stored in a refrigerator for 15 minutes before use in PFGE.
3.5.2. PFGE, Monitoring, and Data Analysis
Here, 1% pulsed-field certified agarose (Bio-Rad, Hercules, CA USA) in 0.5X TBE (Tris/borate/EDTA) buffer (pH 8.4) was casted in a PFGE chamber. Agarose molds treated with XbaI restriction enzyme were then implanted into the wells of the pulsed-field agarose gel using a PFGE comb. The agarose gel implanted with bacterial DNA was then placed into chamber of the PFGE instrument filled with 1900 - 2000 mL of 0.5X TBE buffer. The following PFGE experimental parameters were used: restriction enzyme: FastDigest Xbal; pulse angle: 120°; current: 6 V/cm2; temperature: 14°C; time: 20 hours. Following the PFGE experiment, the agarose gel was transferred into a solution of ethidium bromide (5 µg/mL) in 400 mL of distilled water for staining. The gel was then destained in distilled water.
DNA bands on the destained agarose gel were pictured by GeneGenius gel bio imaging system (Sygene) and saved in TIFF format. DNA band profiles were analyzed by GelCompar II Software version 3.0 (Applied Maths, Sint-Martens-Latern, Belgium). The pictures were normalized based on reference DNA bands from E. coli strain ATCC 25922. Dendogram analysis of the PFGE profiles was conducted by an unweighted pair group method using arithmetic averages (UPGMA) (11). Escherichia coli strains were correlated on PFGE dendogram by the Dice correlation coefficient.
The relatedness of PFGE bands were evaluated based on 4 categories proposed by Tenover et al. (10): Indistinguishable, closely related, possibly related, and unrelated. Here, genetically indistinguishable isolates were epidemiologically identical isolates, referring to the same number of bands with the same size. Closely related isolates differed in a single genetic event; i e, a point mutation or insertion or deletion of DNA, leading to 2 - 3 fragment differences. Possibly related isolates were less likely to be epidemiologically related, involving 2 independent genetic events that gave rise to 4-6 band differences brought by insertions/deletions on DNA or the gain/loss of restriction sites. Isolates with more than 2 genetic events, leading to more than 7 fragments, were considered genetically unrelated.
4. Results
Extended spectrum β-lactamase producing E. coli strains were isolated by culturing 39 urine, 1 tracheal aspiration, 2 wound, 1 sputum, 1 vaginal, and 1 urine catheter samples at the Mevlana University Foundation Hospital (MUFH-Konya) and the Mevlana University Medical Center in Konya (MUMC-Konya) from December 2013 to December 2014 (Figure 1).
An antibiotic resistance profile and rate for 45 ESBL-producing E. coli isolates are given in Figure 1 and Table 1, respectively. As mentioned in the materials and methods section, ESBL-producing E. coli couples 3 - 4 and 9 - 10 (Figure 1) were obtained from 2 different patients. Possibly these isolates were originate from different E. coli strains as they exhibited different antibiotic resistance profiles; for instance, with ampicillin-sulbactam, cefepime, aztreonam, and fosfomycin for isolates 3 - 4 and with ampicillin-sulbactam for isolates 9 - 10. As a result, a total of 45 ESBL-producing E. coli isolates, 9 isolated from 8 inpatients, were considered in all statistical analyses. The reason why the ESBL-producing E. coli species isolated from inpatients were included in the current study was to see if there were any relationships between isolates obtained from inpatients and outpatients. A total of 342 specimens were taken from both inpatients (n = 14) and outpatients(n = 326), overall, the ratio of ESBL-producing E. coli isolates was 13.1%, while this ratio for outpatients was 11% (36 ESBL-producing E. coli isolates out of 326 specimens) and that of inpatients was 64.2% (9 ESBL-producing E. coli isolates out of 14 specimens).
| Antibiotic | R (%) | R + I (%) |
|---|---|---|
| Ampicillin | 100 | - |
| Carbenicillin | 97.8 | - |
| Ampicillin-sulbactam | 57.7 | 66.7 |
| Amoxicillin-clavulonate | 53.3 | 80 |
| Piperacillin | 97.7 | 100 |
| Piperacillin/tazobactam | 22.2 | 35.6 |
| Ceftazidime | 51.1 | 64.4 |
| Cefotaxime | 97.3 | 95.6 |
| Cefoperazone | 97.8 | 100 |
| Cefepime | 24.4 | 28.9 |
| Cefuroxim | 97.8 | 100 |
| Cefixime | 88.9 | 91.1 |
| Cefoxitin | 17.7 | 97.8 |
| Cefaclor | - | 100 |
| Cefazolin | 100 | - |
| Ceftriaxone | 97.8 | 100 |
| Aztreonam | 53.3 | 75.6 |
| Imipenem | 0 | 0 |
| Meropenem | 2.2 | - |
| Tetracyclin | 77.8 | - |
| Trimethoprim-sulfamethoxazole | 88.9 | - |
| Ciprofloxacin | 64.4 | - |
| Netilmicin | 26.7 | - |
| Amikacin | 2.2 | - |
| Gentamycin | 46.7 | 48.9 |
| Nitrofurantoin | 2.2 | 4.4 |
| Fosfomycin | 6.6 | - |
Abbreviations: I, intermediate resistance; R, resistance.
A PFGE dendogram for the 45 E. coli isolates is shown in Figure 1. Based on PFGE analysis criteria proposed by Tenover et al. (10), 4 profiles were epidemiologically different from each other: Profile A that was one of the least commonly identified profiles (2%), profile B that was another least commonly identified profile (2%), profile C that was the most commonly identified profile (67%), and profile D that was the second most commonly identified profile (29%) (Figure 1). Profile C possessed 6 sub-profiles: profiles C1 (7%), C2 (27%), C3 (13%), C4 (2%), C5 (16%), and C6 (2%) (Figure 1). The majority of the ESBL-producing E. coli isolates were resistant to penicillins, cephalosporins, and monobactam (Table 1).
5. Discussion
Resistance of ESBL-producing E. coli strains to antibiotics was a growing problem over the last decade worldwide. The public health institution of Turkey (PHI-Turkey) reported in 2011 that the rate of ESBL-producing E. coli strains was 21% in Turkey (11). The prevalence of ESBL-producing E. coli strains in the USA was 3.9%, 36% of which was due to community-acquired infections from 2009 to 2010 (12). The rate of ESBL-producing E. coli strains in Canada was 5.5 out of 100,000 people, 71% of which was due to community-acquired infections (13). The rate of community-acquired ESBL-producing E. coli infections in Israel was reported 57.8% in 2004 (14), while these rates were 1.5% - 7.6% in Brazil, 3.5% in Italy, and 4.5% in Saudi Arabia (15). The ESBL rate was 13.3% in the studied hospital, which seemed about 8% lower than that of reported by PHI-Turkey and relatively higher than that of the developed Western countries (16).
According to a multi-center antibiotic resistance study in Turkey in 2011, published by the Turkish national antimicrobial resistance surveillance system at PHI-Turkey, ESBL-positive E. coli isolates were resistant to aminopenicillins (75%), aminoglycosides (24%), fluoroquinones (48%), and third generation cephalosporins (44%). Based on the antibiotic resistance rates given in Table 1 for the ESBL-producing E. coli isolates, the antibiotic resistance (R) rate was 100% for aminopenicillins (based on ampicillin data only), 19% for aminoglycosides, 64% for fluoroquinolones (based on ciprofloxacin data only), and 87% for third-generation cephalosporins. As compared to the aforementioned Turkish national data published in 2011 by PHI-Turkey, it seems that resistance of ESBL-producing E. coli strains to aminopenicillins, fluoroquinolones, and third-generation cephalosporins increased significantly in Konya province of Turkey, while resistance to aminoglycosides was about 6% lower than that of reported by PHI-Turkey.
It was determined in the studied laboratory that ESBL-producing E. coli isolates acquired high resistance against cephalosporin antibiotics, confirming that these isolates were ESBL-producing E. coli strains that possibly possessed chromosomal class C (AmpC) ESBL for non-oxyimino cephalosporins (i e; cefoperazone, cefoxitin, cefaclor, and cefazolin) as well as plasmid encoded TEM-, SHV-, or CTX-M-type ESBLs for oxyimino-cephalosporins (i e; cefuroxime, ceftazidime, cefotaxime, and ceftriaxone). In general, community acquired ESBL-producing E. coli isolates evaluated in the studied hospital expressed high susceptibility to carbapenems by 97.8% - 100%, amikacin by 97.8%, nitrofurantoin by 95.6%, and fosfomycin by 93.4%, while mild susceptibility to cefoxitine by 82.3%, netilmicin by 73.3%, and cefepime by 71.1%.
Pulsed-Field Gel Electrophoresis dendogram in Figure 1, determined using the ESBL-producing E. coli isolates, revealed that profiles A, B, C, and D were genetically unrelated by < 85% clonal similarity. Majority of the ESBL-producing E. coli species isolated from outpatients with community-acquired infections in Konya were grouped into profile C family with 6 genetically closely related subprofiles. In regards to ESBL production against cephalosporins in profile C, lines 9, 23, 29, and 34 in the PFGE dendogram in Figure 1 had the same antibiotic profile against cephalosporins, while these ESBL-producing E. coli isolates possess different antibiotic resistance profiles against antibiotics other than cephalosporins. It was found that all subprofiles of profile C were epidemiologically closely related to each other with > 85% clonal similarity, caused likely by a single genetic event leading to 2 - 3 fragment differences. Here, profile C2 was the most commonly identified subprofile (12 specimens out of 45 (27%).
It should be noted here that ESBL-producing isolates marked in lines 37 - 45 in Figure 1 were obtained from inpatients, which all shared the same genotypic profile, profile D. Each of the antibiotic profiles of ESBL-producing E. coli species isolated from inpatients admitted to the intensive care unit (ICU) at MUFH, shown in Figure 1, possessed different antibiotic resistance profiles, suggesting the likelihood that these E. coli strains were rather presented to the hospital by patients, but not produced from nosocomial infections. Likewise, antibiotic profile of E. coli isolates from inpatients in other departments of the hospital including pediatrics service, general surgery service, and orthopedics service possessed different antibiotic profiles; increasing the possibility of community-onset infection (Figure 1). Luckily, cross-contamination between different units of the hospital was not observed. In addition, 4 outpatients with different antibiotic resistance profiles also shared the same genotypic profile D as the outpatients. Differences noticed with the antibiogram of ESBL-producing E. coli isolates possessing the same PFGE profile indicated that the isolates possessed identical chromosomal Xba1 cutting sites with slightly varying genetic sequences between the cutting sites, resulting in slightly different antibiotic resistance profiles.
Interestingly, ESBL-producing E. coli isolates in profile D as well as in subprofiles C2, C3, and C5 were predominantly resistant to penicillins, cephalosporins, and monobactams, while they exhibited a variable resistance profile against aminoglycosides. It is likely that resistance to aminoglycosides was of plasmid origin as the E. coli strains shared the same PFGE bands in the aforementioned profiles.
Prevalence of ESBL-producing E. coli strains in community-acquired infections significantly increased recently in Turkey. The current study laboratory data indicated that majority of the E. coli strains isolated from under study hospitals were highly resistant to penicillins, cephalosporins, and monobactam, posing an imminent threat in the treatment of future community-acquired E. coli infections in Konya. Therefore, it seems crucial to prescribe more susceptible antibiotics against future E. coli infections based on the antibiotic susceptibility data (Figure 1).
6. Conclusion
It was concluded that carbapenems (i e, imipenem and meropenem), amikacin, nitrofurantoin, or fosfomycin can be prescribed as the first-line treatment of future community-acquired E. coli infections, while cefoxitine, netilmicin or cefepime should be considered as the second-line treatment for such infections in Konya province. Due to very low number of samples collected from inpatients, it is too early to conclude the presence of a nosocomial E. coli infection in the studied hospitals from 2013 to 2014.