1. Background
Based on the presence of polysaccharide capsule, Haemophilus influenzae is classified into 2 major groups; typeable H. influenzae and nontypeable H. influenzae (NTHi) (1, 2). The type b strain of typeable H. influenzae (Hib) causes serious invasive infections such as bacteremia, pneumonia, and acute bacterial meningitis (3). The pathogenesis of diseases related to NTHi begins with bacterial colonization of the nasopharynx after spreading the organism into the middle ear, the sinuses, and the lungs resulting in otitis media, sinusitis, bronchitis, and pneumonia (4, 5).
There is no vaccine against NTHi-induced diseases (6). Prevention of NTHi infections could provide considerable health and economic benefits hence efforts are made toward identifying bacterial structures with the potential of being vaccine candidates. Some surface-exposed proteins such as high molecular weight 1(HMW1), high molecular weight (HMW2), H. influenzae type a (Hia), Haemophilus-Actinobacillus-Pasteurella (HAP), P2, P5, and protein D" of NTHi play an important role in adherence to the epithelial cells during colonization (7-9). Among these proteins, HMW1, HMW2, and Hia are the major virulence factors of NTHi. They are high-molecular weight antigens with important roles during the attachment to the surface of epithelial cells in colonization (9-11). The regions that mediate attachment to the epithelial cells named binding domains localized in amino acids 555-914 (HMW1), 553-916 (HMW2), and 585-705 (Hia) of protein adhesins, respectively (12, 13).
2. Objectives
The current study aimed at investigating the potential of recombinant HMW1555-914, HMW2553-916, and Hia585-705 proteins as subunit vaccines to induce immune responses after active immunization in Bagg albino (inbred research mouse strain) or Balb/c mice.
3. Methods
3.1. Ethics Statement
All mouse experiments were performed in accordance with the animal care as well as ethical codes of Tehran University of Medical Science (guideline for working with animals) granted to the current study (93-02-87-26048). The protocol was reviewed and approved by the ethical committee of Tehran University of Medical Sciences, Tehran, Iran.
3.2. Bacterial Strains and Cloning/Expression Vectors
Nontypeable Haemophilus influenzae strain (ATCC 49766) was provided from the Department of Mycobacteriology and Pulmonary, Pasteur Institute of Iran. The strain was cultured at 37°C on chocolate agar, and then, grown in Luria-Bertani broth medium (Merck, Germany). Escherichia coli TOP10 (Novagen, USA) and BL21 (DE3) (Novagen, USA) were respectively used as a cloning and expression system. Cloning and expression vectors were pTG19-T (Vivantis, Malaysia) and pET28a (Novagen, USA), respectively.
3.3. Amplification of HMW1555-914, HMW2553-916 and Hia585-705
Genomic extraction was performed by the DNA extraction kit (DNA Technology, Russia) according to the manufacturer’s instruction. Fragments comprising 1080, 1092, and 363 bp of HMW1555-914, HMW2553-916, and Hia585-705 were respectively amplified by the polymerase chain reaction (PCR) technique (Table 1). PCR amplifications were carried out in a total volume of 20 μL containing 50 ng of genomic DNA, 2 μL of 10X reaction buffer, 0.2 mM of dNTP (Cinnagen, Iran), 2 mM of MgSO4, 0.5 pM of each primer (Bioneer, South Korea), and 1 unit of Pfu DNA polymerase (Thermoscientific, USA). The recovery of PCR product was carried out by a High Pure PCR product purification kit (Vivantis, Malaysia) according to the manufacturer’s instruction.
| Primers | |
|---|---|
| hmw1F: | 5’ -CCA TGG TTG ATG TTC ATA AAA T- 3’ |
| hmw1R: | 5’ -CTC GAG TAC ATT AAA AGT GAA ATT TGT- 3’ |
| hmw2F | 5’ -CCA TGG TTG ATG TTC ATA AAA ATA T- 3’ |
| hmw2R: | 5’ -CTC GAG AAA ATT ACC TGT AAT ATT- 3’ |
| hiaF: | 5’ -CCA TGG ACA TCT CAG CCG GC - 3’ |
| hiaR: | 5’ -CTC GAG GGC CAG TTC AAA CGT- 3’ |
aUnderlined sequences are the restriction enzyme sites.
3.4. Cloning and Subcloning of HMW1555-914, HMW2553-916, and Hia585-705
Retrieved PCR products underwent Taq treatment. In order to add A 3’- overhang, 50 ng DNA, 0.2 mM dATP (Thermoscientific, USA), 1 µL 10X buffer, and 1 unit of Taq polymerase (Cinnage, Iran) were used. The mixture was incubated at 90°C for 10 minutes proceeding to 72°C for 30 minutes. The products were ligated into pTG19-T vector based on the calcium chloride transformation process. The recombinant pTG19-T vectors were digested by NcoI and XhoI (Thermoscientific, USA) and the above mentioned genes were cloned into pET28a. After confirmation with colony-PCR (Table 1), the recom¬binant plasmids were extracted by GF-1 Plasmid DNA extraction kit (Vivantis, Malaysia) and confirmed by DNA sequencing.
3.5. Expression of HMW1555-914, HMW2553-916, and Hia585-705 Proteins
The recombinant plasmids were transformed into E. coli BL21 (DE3). The expression of proteins was induced by adding 1 mM of isopropyl-β-D-thiogalactopyranoside (IPTG) (Cinnagen, Iran). After 4 hours induction, the cells were harvested at the optical density (OD) of 2 at 600 nm. The pellets of 100 mL culture medium were resuspended in 10 mL of lysis buffer (300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, and pH: 8). All reagents were from Sigma, USA. Then, the suspension process was disrupted by sonication (5 minutes, 50% of amplitude, comprising of 30 seconds on, 30 seconds off) on ice. The proteins were analyzed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). To run SDS-PAGE, the bacterial pellets were suspended in loading buffer, heated to 90°C for 10 minutes.
3.6. Purification of Recombinant HMW1555-914, HMW2553-916, and Hia585-705 Proteins
The pellets of culture medium (Merck, Germany) were resuspended in lysis buffer. Under denaturating conditions, the proteins were purified by affinity chromatography on Ni-nitrilotriacetic acid (Ni-NTA) (Qiagen, Netherlands) based on the manufacturer’s instruction. The proteins loaded on a column were pre-equilibrated with denaturing binding buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH: 8). In order to refold the target proteins, the columns were subjected to a linear gradient of urea from 8 to 0 M in the refolding buffer (20 mM sodium phosphate, 300 mM NaCl, and pH: 7.2; all from Sigma, USA). The contaminant proteins were washed away by the washing buffer (20 mM sodium phosphate, 300 mM NaCl, 20 mM imidazole, pH 8.0; all from Sigma, USA). Finally, target proteins were eluted by the elution buffer (20 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole, pH: 8). The eluted target proteins were concentrated by Amicon filters (Millipore, USA).
3.7. Western Blot Analysis
The Western blot analysis was applied following SDS-PAGE, and then, the target proteins were transferred to polyvinylidene difluoride (PVDF) membrane (Hi-bond Amersham Biosciences, USA) by the wet system (Bio-Rad, USA). The membranes were blocked with 2% (w/v) BSA (Sigma, USA) in phosphate buffer saline (PBS), and then, washed with PBST (PBS 1% + Tween 20). The membrane was incubated with anti-His tag antibody conjugated with horseradish peroxidase (HRP) (Abcam, Canada) (1:3000 diluted in PBS) at room temperature. The presence of recombinant HMW1555-914, HMW2553-916, and Hia585-705 proteins were analyzed by adding diaminobenzidine (DAB) (Roche, Germany) solution.
3.8. Mouse Immunization
Groups of 6-week-old BALB/c female mice were immunized with purified proteins alone, or in binary and ternary combinations. The immunization protocol consisted of 3 subcutaneous injections of 100 µg purified proteins mixed with the equal volume of adjuvant administered every 2 weeks. The first dose of injection was mixed with the Freund complete adjuvant, and the following doses were injected with incomplete the Freund adjuvant. The control group was immunized with PBS. Optimum dilution (1:128) was determined prior to the comparisons, by testing serially diluted sera against the coated antigens.
3.9. Determination of IgG levels against HMW1555-914, HMW2553-916, and Hia585-705
Total IgG (the days 0, 14, and 28) and IgG isotypes responses (IgG1 and IgG2a) (day 28) of the immunized mice were evaluated by ELISA technique. The IgG antibody responses of Balb/c mice against purified HMW1555-914, HMW2553-916, and Hia585-705 proteins were determined by ELISA. Serum samples were collected at weeks 2 and 4 after the first injection. Briefly, 96-well flat-bottomed plates (Falcon, USA) coated by 1µg purified proteins were incubated overnight at 4°C. Purified proteins were used in PBS buffer (pH: 7.4) at a concentration of 10 µg/mL, and then, the plates were blocked with 5% (w/v) BSA for 2 hours at room temperature; thereafter, it was washed by PBS-T and then, incubated with antisera (1: 128) for 2 hours at room temperature. The wells were incubated by goat anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad, Hercules, Cal, USA) with dilution of 1:3000. After 2 hours of incubation at room temperature and several washings with PBS-T, the wells were incubated with 100 μL tetramethyl benzydine (TMB), as a substrate solution, for 15 minutes at room temperature. Reaction was stopped by adding H2SO4 2N and the absorbance was measured at 450 nm by the ELISA Reader (Biotek, USA).
3.10. Serum Bactericidal Assay
Serum bactericidal assay was applied as previously described (14). After overnight incubation of NTHi strain on chocolate agar, single bacterial colonies were inoculated into Luria-Bertani broth medium. The bacteria were grown for 5 hours to enter the log phase. After incubation, they were pelleted by centrifugation, resuspended in PBS containing 9 mM CaCl2, 4.9 mM MgCl2, and 1% (w/v) BSA (the Dulbecco buffer). In the current study, the SBA was performed on pooled serum specimens collected at weeks 0 and 4, and all sera were inactivated by heating for 30 minutes at 56°C. As an active source of exogenous complement, a four-week-old baby rabbit serum was used (10% v/v). To measure functional activity, sterile 96-well flat-bottom plates were used. The mixture contained 25 µL complement, 50 µL diluted sera, and 25 µL buffer containing 500 CFU of bacteria. After addition of mixture to each well, the pellet was incubated for 1 hour at 37°C in a 5% CO2 and then, a sample of each well (5 µL) was plated onto a chocolate agar and incubated for 16 hours at 37°C. All assays were performed in duplicate.
3.11. Statistical Analysis
All statistical analyses were performed using SPSS 18.0 (SPSS, Chicago, IL, USA). Due to the normalization of the data, one-way ANOVA and t test were used to evaluate the differences between data sets. P value < 0.05 was considered statistically significant.
4. Results
4.1. Construction of Recombinant Vectors
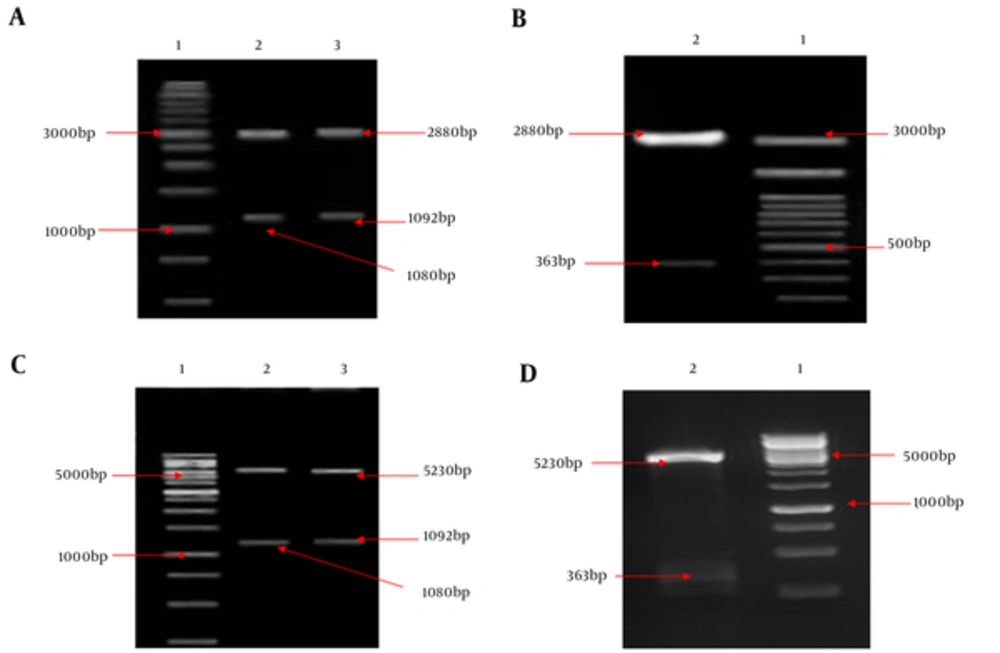
HMW1555-914, HMW2553-916, and Hia585-705, as binding domains, play an important role in the colonization of NTHi in the nasopharynx during infections. Binding domains of adhesion proteins have a relatively conserved sequence and are exposed at surface of outer membrane among NTHi strains. Coding sequence fragments of HMW1555-914 (1080 bp), HMW2553-916 (1092 bp), and Hia585-705 (363 bp) were cloned into pTG19 and pET28a, respectively. Recombinant vectors were confirmed by double digestion (Figure 1) and sequencing.
A, Lane 1: 1 kb DNA size marker, lane 2: Double digestion of the recombinant pTG19/HMW1555-914 (pTG19 backbone: 2880 bp and HMW1555-914: 1080 bp), lane 3: Double digestion of the recombinant pTG19/HMW2553-916 (pTG19 backbone: 2880 bp and HMW2553-916: 1092 bp); B, Lane 1: 100 bp DNA size marker, lane 2: Double digestion of the recombinant pTG19/Hia585-705 (pTG19 backbone: 2880 bp and Hia585-705: 363 bp); C, Lane 1: 1 kb DNA size marker, lane 2: Double digestion of the pET28a/HMW1555-914 (pET28a backbone: 5230 bp and HMW1555-914: 1080 bp), lane 3: Double digestion of the pET28a/HMW2553-916 (pET28a backbone: 5230 bp and HMW2553-916: 1092 bp); D, Lane 1: 1 kb DNA size marker, lane 2: #: HMW1555-914, HMW2553-916 and Hia585-705 as Su... Revision 1 Journal: Jundishapur Journal o... Page 22 of 28 5 August 2017 05:10:58 Double digestion of the pET28a/Hia585-705 (pET28a backbone: 5230 bp and Hia585-705: 363 bp)
4.2. Expression and Purification of HMW1555-914 and HMW2553-916 proteins
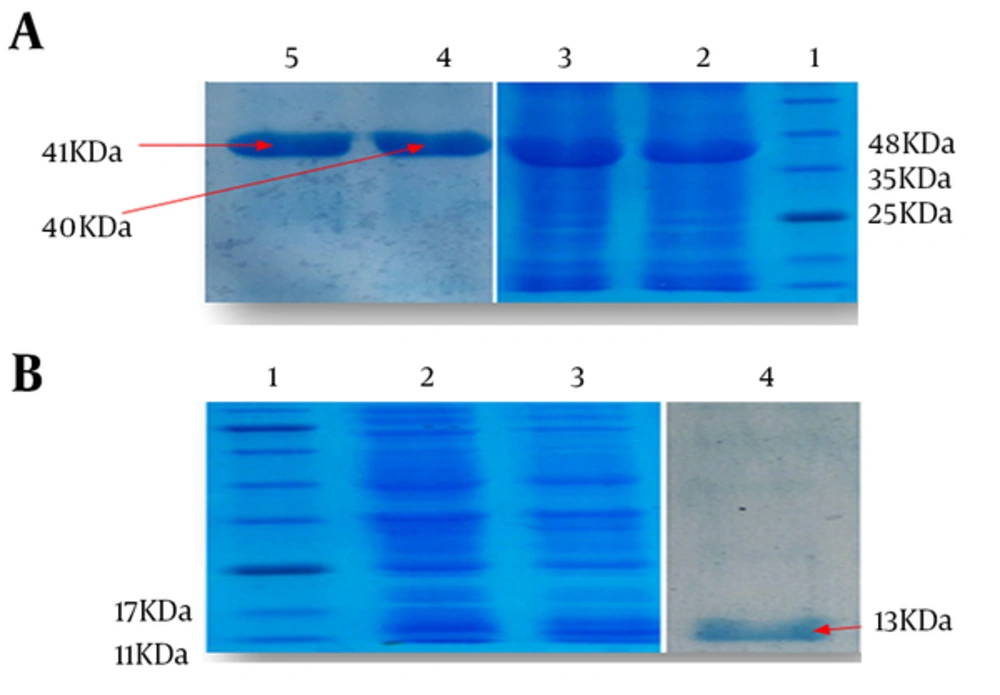
Strain of E. coli BL21 (DE3) harboring pET28a/HMW1555-914, pET28a/HMW2553-916, and pET28a/Hia585-705 were optimized to achieve high levels of protein expression after 4 hours of induction. Recombinant expressed proteins of HMW1555-914, HMW2553-916, and Hia585-705 were mainly expressed in the form of inclusion body carefully purified under denaturing condition by Ni-NTA. Major bands of the proteins appeared approximately at the size of 40, 41, and 13 kDa expected according to the molecular weight of HMW1555-914, HMW2553-916, and Hia585-705, respectively (Figure 2).
A, Lane 1: standard protein size marker, Lane 2: Induced expression of HMW1555- 914 with molecular weight of 40 kDa, Lane 3: Expression of HMW2553-916 with molecular weight of 41 kDa, Lane 4: purified HMW1555-914, Lane 5: purified HMW2553-916; B, Lane 1: standard protein size marker, Lane 2,3: induced expression of Hia585-705 with molecular weight of 13 kDa, Lane 4: purified protein of Hia585-705
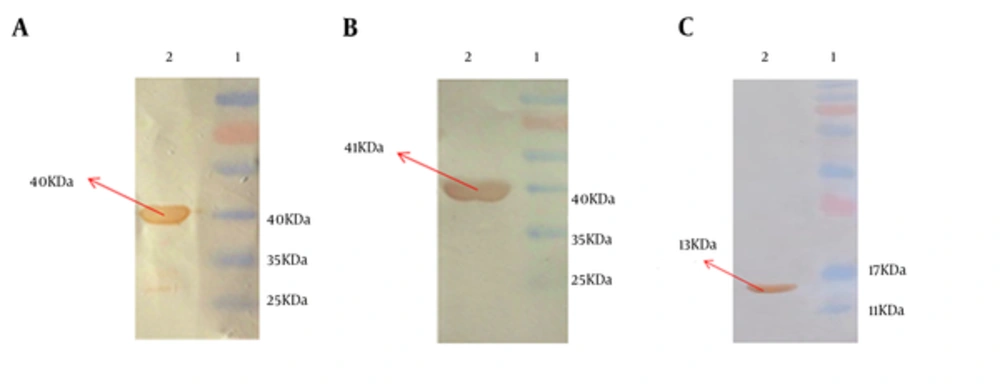
4.3. Western Blot Analysis
Recombinant purified HMW1555-914, HMW2553-916, and Hia585-705 proteins were observed at the size of 40, 41, and 13 kDa, respectively. The identity of recombinant proteins was approved by anti-His tag monoclonal antibody (Figure 3).
4.4. IgG specific Antibody Responses after Mouse Immunization
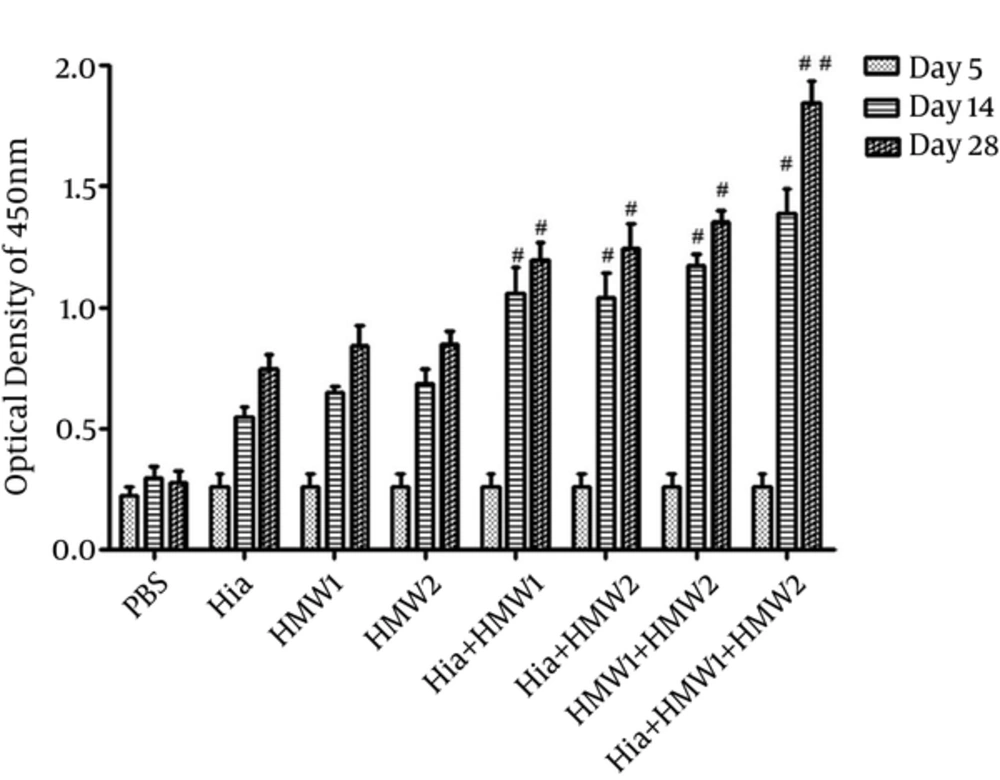
Mice (Balb/c) immunized subcutaneously with recombinant HMW1555-914 in combination with HMW2553-916, and Hia585-705 proteins showed the highest levels of specific antibody responses (Figure 4). IgG responses stimulated with HMW1555-914 in combination with HMW2553-916, and Hia585-705 (ternary combination) were significantly higher than the ones induced by HMW1555-914, HMW2553-916, and Hia585-705 alone and in binary combinations (Hia + HMW1, Hia + HMW2, and HMW1 + HMW2) (Figure 4).
Although there was significant increase in IgG antibody responses in binary combination (Hia + HMW1, Hia + HMW2 and HMW1 + HMW2) in comparison to the groups that received each of HMW1555-914, HMW2553-916, and Hia585-705 alone, these responses were low compared with the group immunized with ternary combination (Hia + HMW1 + HMW2) (Figure 4). The booster injections (especially the second booster) immunized with HMW1555-914, HMW2553-916, and Hia585-705 in binary and ternary combinations were significantly effective to increase anti-HMW1555-914, HMW2553-916, and Hia585-705 IgG responses in comparison to the control group. The response to the second booster was statistically higher than that of the first booster (P ≤ 0.05) in ternary combination immunized with HMW1555-914 in combination with HMW2553-916 and Hia585-705 (Figure 4). Groups immunized solely with HMW1555-914, HMW2553-916, and Hia585-705 showed antibody rises, but the increases were statistically insignificant (P ≥ 0.05) (Figure 4).
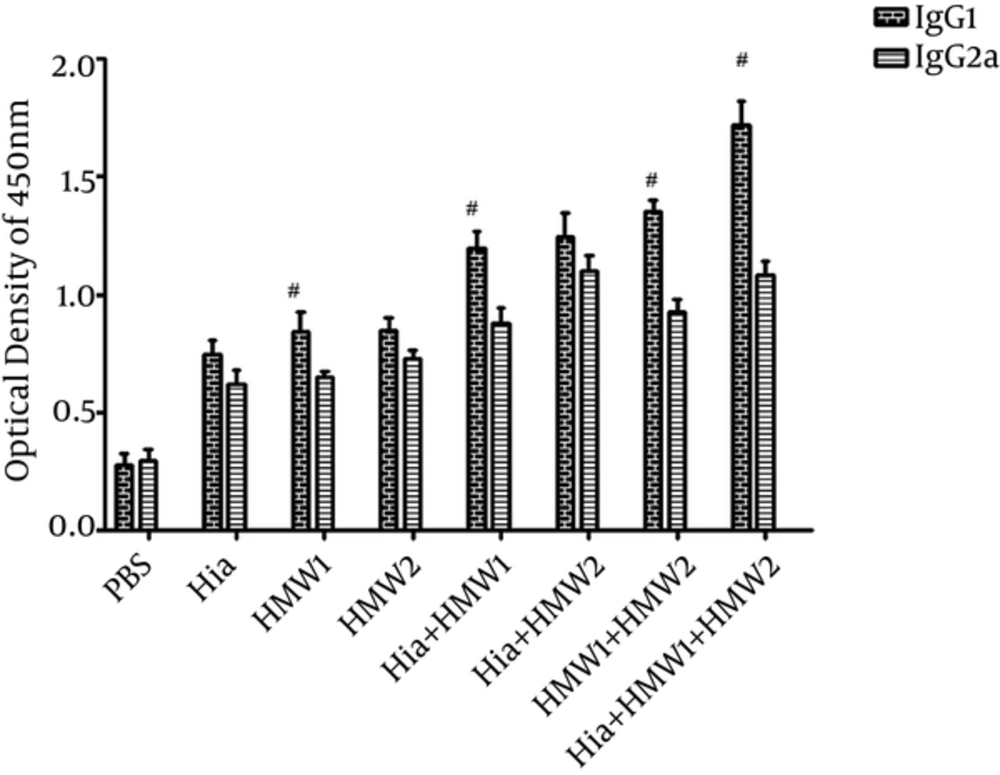
To assess IgG1 and IgG2a levels, antisera were collected 1 week after the second booster. Antibody responses against each of HMW1555-914, HMW2553-916, and Hia585-705 while used alone or in binary or ternary combinations were mostly composed of IgG1 other than IgG2a (Figure 5). In Balb/c mice immunized by HMW1555-914 alone or in binary (Hia + HMW1 and HMW1+ HMW2) and ternary (Hia + HMW1 + HMW2) combinations, IgG1 was significantly higher than IgG2a (P ≤ 0.05). Mice (Balb/c) immunized by ternary combination compared with the groups immunized solely by HMW1555-914, HMW2553-916, and Hia585-705, and in binary combinations, showed significantly higher responses of IgG1 than IgG2a (P ≤ 0.05) (Figure 5).
4.5. Serum Bactericidal Assay
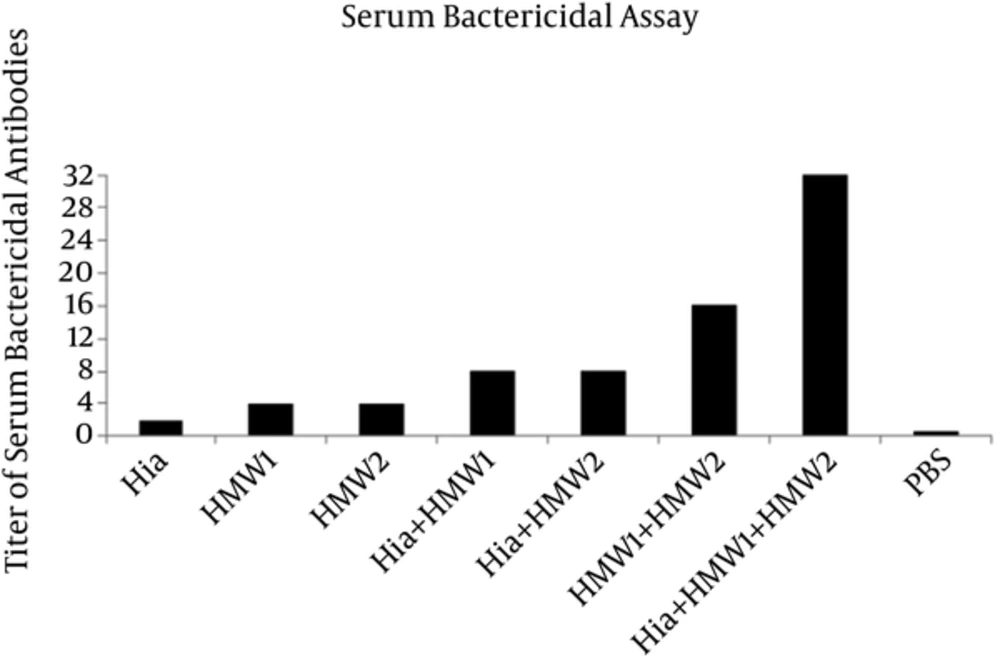
The antisera were tested for the in vitro ability to promote killing of NTHi strain (ATCC 49766) by complement system. The sera from animals immunized by HMW1555-914 in combination with HMW2553-916 and Hia585-705 showed the highest bactericidal activity against NTHi strain with the titer of 1:32 (Figure 6). The antisera of the mice immunized by Hia + HMW1, Hia + HMW2, and HMW1 + HMW2 showed less bactericidal activity in comparison with that of ternary combination with titers of 1:8, 1:8 and 1:16, respectively (Figure 6). Exclusive usage of Hia, HMW1, and HMW2 elicited less bactericidal activity with titers of 1:2, 1:4 and 1:4 in comparison with those of the binary and ternary combinations of such domains (Figure 6).
5. Discussion
By preventing the colonization of NTHi, otitis media, pneumonia, and sinusitis caused by this bacteria dramatically decreased (15). There is no vaccine against NTHi-induced diseases (6). HMW1, HMW2, and Hia are the protein adhesins that mediate attachment to a variety of pharyngeal epithelial cells. HMW1, HMW2, and Hia are identified as major targets of the human antibody responses (11). The regions of HMW1, HMW2, and Hia proteins interacting with the epithelial cells are called binding domains. These regions are localized in 555 - 914th, 553 - 916th, and 585 - 705th amino acids in HMW1, HMW2, and Hia, respectively (12, 13). These protein adhesins could be good vaccine candidates due to their important roles in protection against NTHi infections (9). Based on a literature review, it is for the first time in the world that the induction of specific antibody responses by HMW1555-914, HMW2553-916, and Hia585-705 binding domains alone or in binary and ternary combinations is evaluated. It should be noted that binding domains of protein adhesins are composed of the conserved regions of HMW1, HMW2 and Hia. The current study hypothesized that HMW1555-914 in combination with HMW2553-916 and Hia585-705 can boost immune responses and cause higher production of specific antibodies.
Immunization of Balb/c mice by ternary combination of these domains in comparison with using them alone or in binary combinations resulted in a significant increase in IgG specific antibody responses. In addition, ternary combination of domains (Hia + HMW1 + HMW2) could induce more specific antibody response due to their synergistic effect. These specific antibodies had more bactericidal activities against NTHi (ATCC 49766) strain with titer of 1:32. SBA confirmed that antibodies were functional to kill NTHi strain by the complement system. These results were consistent with those of Linda et al. in which specific antibodies directed against native protein adhesins such as HMW1/HMW2 and recombinant Hia proteins could kill homologous NTHi strains. The differences between the results of the current study and those of Linda et al. can be attributed to the protein adhesins. The current study used recombinant binding domains of HMW1/HMW2, while in the study by Linda et al. whole native proteins of HMW1/HMW2 were used (11).
Winter et al. constructed recombinant adenovirus vectors to express the HMW1, HMW2, and Hia protein adhesins of NTHi. Chinchilla models immunized by these DNA vaccines showed specific antibody responses (9). The current study used binding domains of purified proteins to induce antibody responses in Balb/c.
Previous studies showed that HMW1, HMW2, and Hia protein adhesins were the immunogenic proteins and also good vaccine candidates to produce bactericidal antibodies and were previously used to develop vaccine in different ways (9, 11, 16, 17). In the current study, the relative ratios of IgG1 to IgG2a revealed significantly higher IgG1 and IgG2a production in Balb/c mice immunized by ternary combination. However, IgG1/IgG2a ratio indicated that the combination of proteins directed immune responses toward T-helper 2 (Th2) versus T-helper 1 (Th1) cell responses. However, it should be noted that these antibody responses were the good indicators showing the capability of the subunit vaccine candidates. Opsonophagocytic killing assays and neutralization tests showed that HMW1555-914, HMW2553-916, and Hia585-705 protein adhesins had more potency to be used as subunit vaccine candidates instead of whole native proteins.
6. Conclusion
In conclusion, the current study demonstrated that serum IgG responses against purified HMW1555-914, HMW2553-916, and Hia585-705 mediate in vitro killing of NTHi strain (ATCC 49766) by the complement system. Further investigations on HMW1555-914, HMW2553-916, and Hia585-705 protein adhesins, as subunit vaccine candidate for NTHi-induced diseases, are highly recommended.