1. Background
Streptococcus pyogenes (group A streptococcus, GAS) is an important pathogen associated with a range of infections in humans (1). This organism causes common diseases such as pharyngitis, erysipelas, and impetigo, but sometimes can also cause severe invasive diseases such as toxic shock-like syndrome and necrotizing fasciitis (2, 3). Currently, antimicrobial therapy is the first choice for the treatment of GAS; however, antimicrobial resistance is a common problem worldwide and the consumption of antimicrobials has been identified as a driving force for the emergence and transmission of antimicrobial resistance in GAS (4). Streptococcus pyogenes has very complex virulence factor (5). The cell wall of S.pyogenes is associated with a range of proteins, a large number of which are encoded by the emm gene family: M proteins are the members that contain the most important structures for virulence (5-7).
Other cell wall proteins in S.pyogenes include streptococcal collagen-like surface protein (SclA) (8). SclA is transcriptionally regulated by Mga and has a highly conserved signal sequence; in a previous study, SclA was found in 12 S.pyogenes strains tested by PCR (9). Since the M protein is a major virulence factor of GAS, it could be a candidate for use in vaccine design (10). However, previous studies have shown that some epitopes of M proteins can induce autoimmunity and cross-react with human tissues (9, 10), because of which they may not be suitable for use in vaccine design.
2. Objectives
In this study, we characterized the SclA protein of S.pyogenes, using online and off-line computational tools for the development of new vaccines to prevent and treat pyogenesis.
3. Materials and Methods
3.1. Retrieval of the Target Sequence
The candidate organism for the present study was S. pyogenes. The protein sequence of SclA (GenBank: CAC33784.1) was obtained from NCBI (http://www.ncbi.nlm.nih.gov), and a BLAST alignment was performed against Protein Data Bank (PDB) entries to find similar sequences.
3.2. Functional Characterization of SclA
Functional characterization of the SclA protein sequence was performed using Uniprot (http:// expasy.org). Primary structure prediction was computed using the ExPASy ProtParam server (http://expasy.org/cgi-bin/protpraram).
3.3. Protein Topology Prediction and 3D Structure of SclA
The secondary structure of the SclA protein was obtained using the PSIPRED Protein Sequence Analysis Workbench (http://bioinf.cs.ucl.ac.uk/psipred/). Using the TMHMM data bank, we predicted transmembrane helices based on a hidden Markov model and predicted the locations of transmembrane intracellular and extracellular regions. Garnier- Osguthorpe- Robson (GOR) IV was used to predict alpha helices, beta sheets, and random coil secondary structures. The three-dimensional structure of SclA was obtained using http://www.ncbi.nlm.nih.gov/Structure/VAST.
3.4. B Cell Epitope Prediction
The immune epitope database analysis resource (IEDB-AR, http://tools.immuneepitope.org) provides a collection of web-based tools for the prediction and analysis of immune epitopes. In this study, IEDB was used to predict the regions of SclA that are likely to be recognized as epitopes in the context of a B response. Conservancy analysis for epitopes and the percentages of protein sequence matches at identity ≥ 100% were also calculated.
4. Results
In silico analysis can be used for identifying drug targets, designing vaccines, and choosing a rapid strategy for disease control. The sequence of the SclA protein was obtained from the NCBI protein sequence database and a BLAST search was performed against Protein Data Bank (PDB) entries for sequence similarity. SclA was found to be associated with 3HQV_B, a protein from Rattus norvegicus. The score for protein alignment between SclA and 3HQV_B was 30.0, with a 46% identity. PROSITE analysis showed that SclA was a surface-localized protein. Primary structure prediction showed that SclA had 123 amino acid residues and an estimated molecular weight of 13137.6. The total number of positively charged residues (Arg + Lys) was 22 and the total number of negatively charged residues (Asp + Glu) was 21. The very high aliphatic index (48.54) and instability index (36.76) provided an estimate of the stability of protein in a test tube. The Grand Average Hydropathicity (GRAVY) value was -0.956.
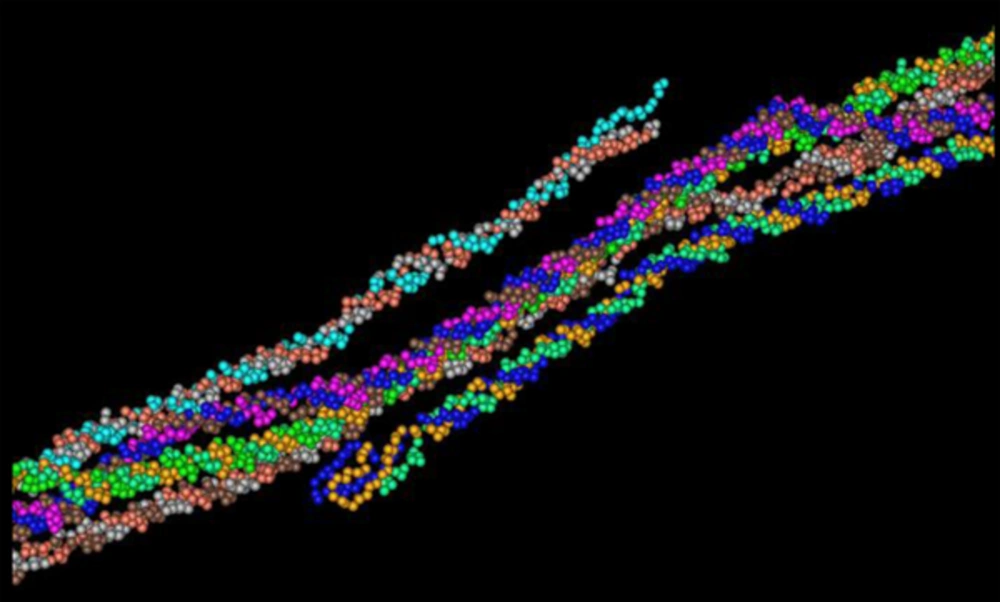
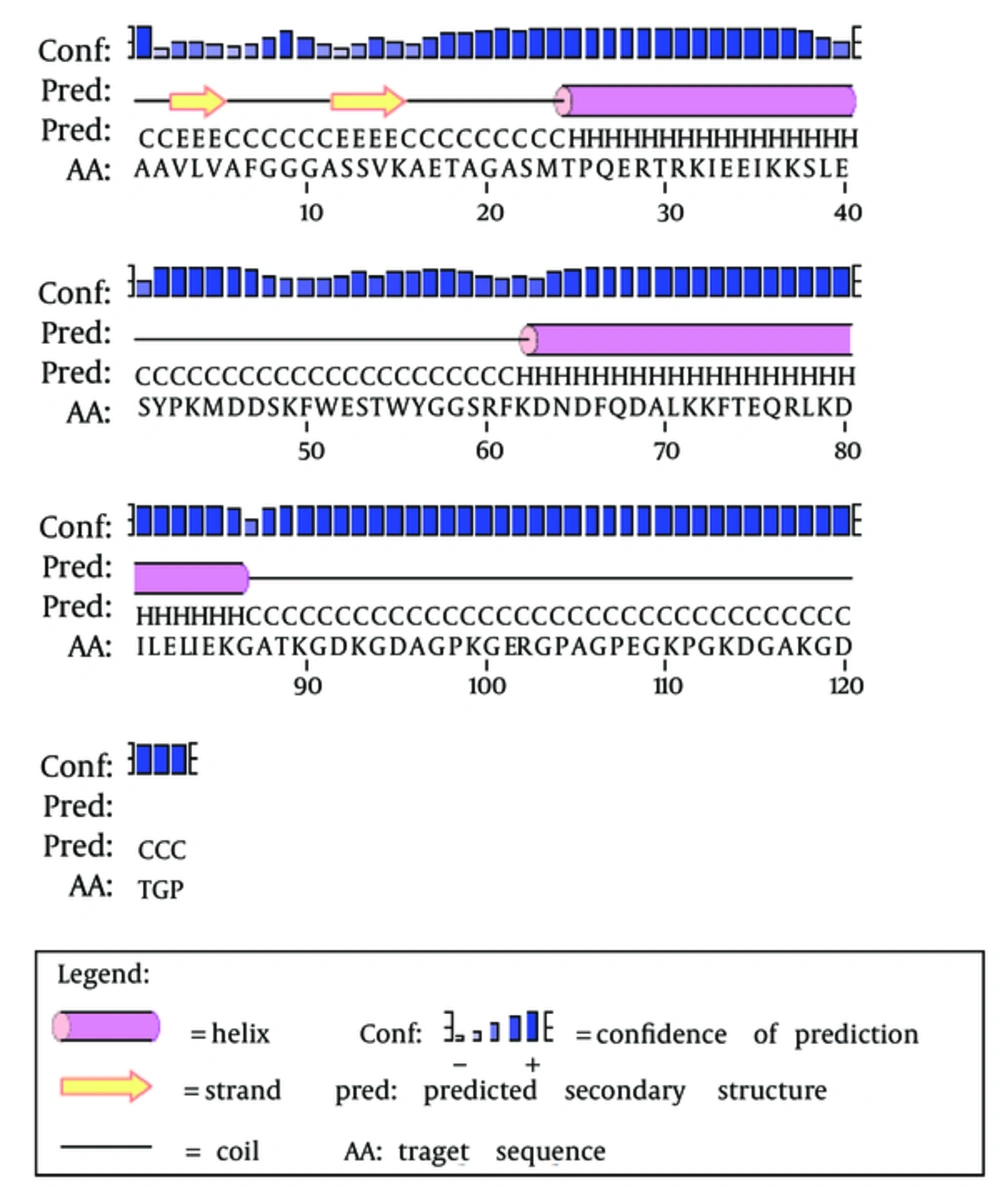
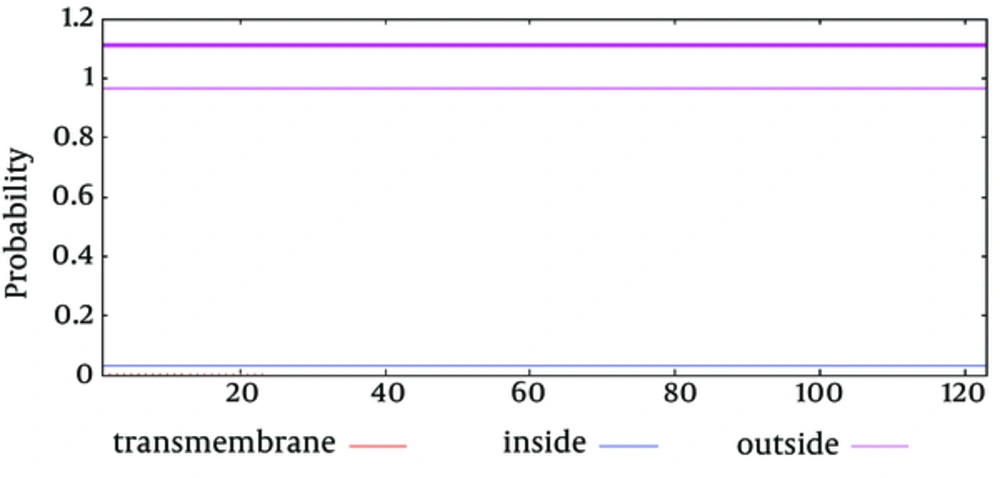
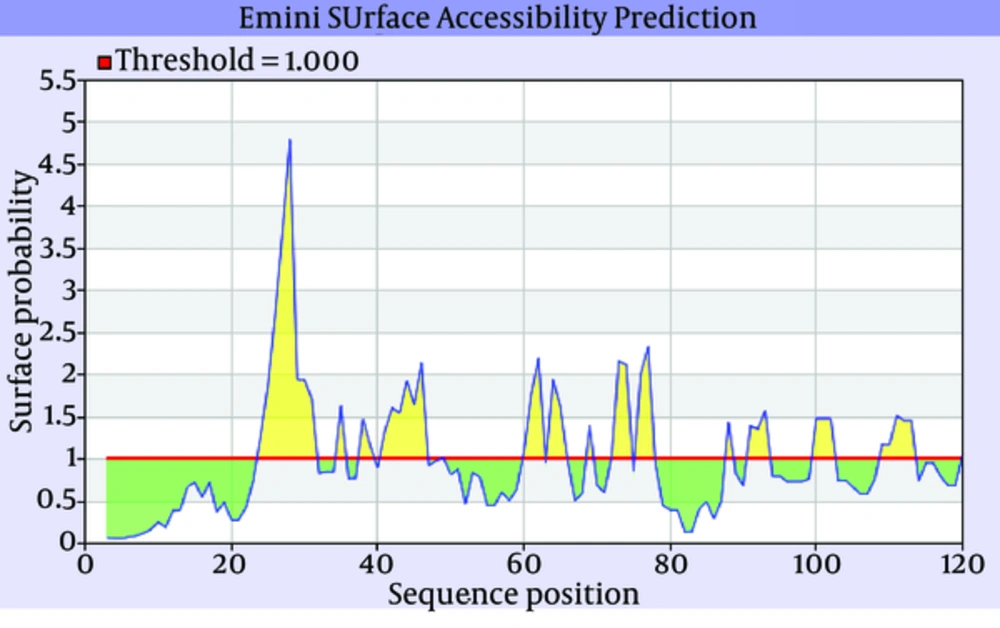
The secondary structure prediction showed that the extended strand (10.57%) and alpha helix (36.59%) were the least frequent, whereas the random coil was more frequent at 52.85%. As the 3D structure of SclA had not been reported in Protein Data Bank (PDB), a BLASTp similarity search was performed against the PDB database and 3HQV_B, where a complete query coverage was obtained (Figure 1). By using PSIPRED, we showed the location and spatial arrangement of each amino acid separately (Figure 2). TMHMM predicted the locations of transmembrane intracellular and extracellular regions and showed that the SclA protein was present on the outside of the cell (Figure 3). The results of the IEDB showed that the maximum length of a peptide that is likely to be predicted as an epitope was of 6 amino acids between positions 26 and 31, with an epitope sequence of QERTRK and a score of 4.791 (Figure 4).
5. Discussion
Streptococcus pyogenes is one of the most pathogenic bacteria infecting children and adolescents. GAS is involved with a wide range of infections and diseases that are important causes of morbidity and mortality, primarily in developing countries, causing > 500,000 deaths annually (2, 11). It is therefore important to treat and prevent these diseases with drugs and vaccines. Currently, antimicrobial therapy is the first choice for the treatment of GAS; however, the emergence of antimicrobial resistance and side effects of antibacterial drugs is increasing (12), resulting in an increased demand for novel drug targets and vaccine design. On the surface of S. pyogenes, 79 surface-localized proteins were reconnoitered and each of them was identified in one growth phase. For example, the expression of SclA was consistent with a previously published report showing that SclA was transcribed in the mid-log phase (13).
The SclA protein contains a signal sequence, a cell wall anchor containing the LPXTGX motif, and proline-rich repeats in the COOH-terminal part; the hypervariable NH2-terminal regions of SclA show that conserved secondary structures interact with a common ligand and have common functions (14). For drug target and vaccine design against many bacterial pathogens, conserved protein sequences should be used. However, vaccine protection was demonstrated to be variable between different strains of the same serotype and different genotypes (15). Unfortunately, an effective vaccine to prevent GAS infection is yet to be developed. Secreted or surface proteins or lipoproteins are favored candidates for vaccines because they are the most easily accessible microbial antigens to the immune system (14, 16). As SclA is a surface protein, it may be a suitable candidate for vaccine design (13).
The focus of the current study was therefore to characterize the SclA protein of S. pyogenes using computational tools. SclA had 123 amino acid residues with a molecular weight 13137.6. The total number of positively charged residues was more than that of negatively charged residues. The N-terminal sequence is considered to contain Alanin (Ala). Garnier- Osguthorpe- Robson (GOR) IV comparative analysis showed that a random coil is predominantly present, followed by an extended strand and an alpha helix. TMHMM determined that the SclA protein is present on the outside of the cell. The results of IEDB showed that the maximum length of the peptide that is likely to be predicted as an epitope is of 6 amino acids, from amino acid 26 to 31, with a score of 4.791 and a protein sequence identity match of 100.00% (1/1). These epitopes can be considered for antibady and drug design.
The information on the characterization of the SclA protein of S.pyogenes obtained from these studies alone is not sufficient. Nonetheless, this information suggests that SclA could be a candidate for more rigorous studies on drug and vaccine design against S.pyogenes.