1. Background
Urinary tract infections (UTI) are the most common bacterial infections worldwide, affecting 150 million people every year and can be involved in any structure of the urinary system such as the urethra, bladder, ureters, and kidney. They are considered the Major cause of ≈ 10 million healthcare visits each year (1). Both genders might be affected by UTI; however, the female population is the most susceptible to be infected due to the closeness of anal and vaginal sites that can result in at least one UTI episode during their lives (2). The epidemiological data for 2016 by the Mexican Health Department showed that the female gender is the main population affected by urinary tract infections (3). These infections caused serious consequences such as high morbidity in a vulnerable population, pyelonephritis, renal damage, preterm birth, and complications by irrational use of antimicrobials (4).
High prevalence, recurrence, and severe health effects resulted in high costs of treatment; therefore, UTI is considered a public health problem (5). Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Enterococcus faecalis, and Staphylococcus saprophyticus are the main pathogens causing UTI (6, 7). Moreover, uropathogenic E. coli (UPEC) has been responsible for more than 85% of UTIs (8). Uropathogenic E. coli pathogenesis begins with periurethral contamination, followed by urethral colonization and subsequent bacterial migration to the bladder (1). Several virulence factors e.g. pili associated-adhesins, including FIC, P, S, type I pili, and PapC have been recognized as receptors in the bladder epithelium and are involved in the colonization event (9-17). Also, host cell colonization and invasion are mediated by the FimH adhesin, localized in the tip of type I fimbriae, due to its capability to bind to mannosylated receptors in the bladder epithelium (18).
Moreover, UPEC synthetizes the cytotoxic necrotizing factor type 1 (CNF1) to increase its capability to access deeper tissue and to persist in the lower urinary tract (19). In recent years, the effectiveness of UTI treatment has become difficult due to the emergence of multidrug-resistant pathogens. In fact, the wide-spectrum β-lactamases (ESBL) are responsible for multidrug-resistance in a large number of genera and species of the Enterobacteriaceae family. Uropathogenic E. coli carries blaSHV and blaTEM genes that are involved in antimicrobial resistance and have been mainly reported as conjugative plasmids (20). Also, blaCTX-M gene encodes an enzyme that hydrolyzes cefotaxime and ceftazidime, and its importance in pathogen resistance has been demonstrated (21). Furthermore, E. coli can produce a carbapenemase (encoded in blaOXA gene) allowing bacteria to confer resistance to carbapenems (22).
2. Objectives
This study was conducted in order to know the genetic distribution and relation of virulence and antimicrobial resistance genes in E. coli strain. In other words, the aim of this work was to identify virulence and resistance genes in UPEC and CEC isolated from patients with UTI in hospitalized and ambulatory patients in Mexico.
3. Methods
3.1. Strains Origin, Identification, and Antimicrobial Resistance Assay
One hundred seven E. coli strains were isolated from ambulatory and hospitalized diagnosed patients with UTI at the “Hospital Juárez de México” (HJM), from April 2016 to January 2017. Urine samples (obtained in the second urine streams) were streaked on Mac Conkey agar and were incubated aerobically at 37°C for 24 - 48 hours. Typical colonies of E. coli were selected and purified on LB agar and were cultured in LB-broth and frozen in glycerol at -70°C. The identification of strains and the antimicrobial susceptibility were performed by using BD Phoenix™ automated identification and susceptibility testing system according to the manufacturer’s protocols.
Antimicrobial susceptibility was performed for 16 antimicrobial agents (according to the microorganism identified “Gram-negative”), by using amoxicillin/clavulanic acid (AMC, 20/10 µg), ampicillin (AM, 10 µg), cefoxitin (FOX, 30 µg), piperacillin/tazobactam (TZP, 100/10 µg), ertapenem (ETP, 10 µg), imipenem (IPM, 10 µg), meropenem (MEM, 10 µg), cefazolin (CZ, 30 µg), cefepime (FEP, 30 µg), cefotaxime (CTX, 30 µg), ceftriaxone (CRO, 30 µg), amikacin (AN, 30 µg), gentamicin (GM, 10 µg), levofloxacin (LVX, 5 µg), nitrofurantoin (FM, 300 µg), and trimethoprim/sulfamethoxazole (SXT, 23.75/1.25 µg). Antibiotic resistance was calculated and represented in percentage (%). Antimicrobial resistance to different antibiotics was confirmed using the disk diffusion method on Mueller-Hilton agar plates according to the guidelines set by “The Clinical and Laboratory Standards Institute (CLSI 100-S21)”. Escherichia coli ATCC 25922 was used as the control.
3.2. Bacterial Phylogenetic Assignment
All the amplification reactions were performed in a SEEAMP™ PCR system (Seegene Inc. Korea). DNA was extracted by using the Qiagen Mini kit (Qiagen, Courtaboeuf, France). PCR reactions were carried out in a total volume of 20 µL with 200 ng of DNA, 10 pmol of each primers pair, 10 mM dNTP’s, 1.5 mM MgCl2, 1X PCR Taq buffer, and 0.25 U of Taq DNA polymerase (Invitrogen). Phylogenetic assignment was performed by using the protocol described by Clermont et al., with modifications (23, 24). The assignment of the criteria for the E. coli phylogenetic analysis was carried out by the amplification of 4 genes (chuA, yjaA, TSPE4.C2, and arpA) corresponding to one of the four phylogenetic groups (A, B1, B2, and D, respectively). Primers for PCR amplification assays are shown in Table 1. Specificity of the primers, a bioinformatic analysis was performed by using MFEprimer-2.0 server available at http://biocompute.bmi.ac.cn/CZlab/MFEprimer-2.0 (25). PCR multiplex conditions for gene fragments were performed as follows: 4 minutes at 94°C followed by up to 30 cycles of 94°C for 5 seconds, 59°C for 20 seconds, 72°C for 1 minutes, and finally, 72°C for 5 minutes. E. coli LMM36-ULA (chuA+, yjaA+) and LMM32-ULA (TSPE4.C2+) strains were used as positive controls.
| Gene | Category | Sequencea | Size, bp | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Phylogenetic assignment | ||||
| chuA | Hemin uptake system | 5’-TGCCGCCAGTACCAAAGACA-3’ | 5’-GACGAACCAACGGTCAGGAT-3’ | 279 |
| yjaA | Unknow | 5’-TGAAGTGTCAGGAGACGCTG-3’ | 5’-ATGGAGAATGCGTTCCTCAAC-3’ | 211 |
| TSPE4.C2 | Anonymous DNA fragment | 5’-GAGTAATGTCGGGGCATTCA-3’ | 5’-CGCGCCAACAAAGTATTACG-3’ | 152 |
| arpA | Ankyrin-like regulatory protein | 5’-AACGCTATTCGCCAGCTTGC-3’ | 5’-TCTCCCCATACCGTACGCTA-3’ | 400 |
| Virulence factors | ||||
| traT | Serum resistance associated | 5’-GGTGTGGTGCGATGAGCACAG-3’ | 5’-CACGGTTCAGCCATCCCTGAG-3’ | 290 |
| fimH | Adhesin | 5’-TCGAGAACGGATAAGCCGTGG-3’ | 5’-GCAGTCACCTGCCCTCCGGTA-3’ | 508 |
| iutA | Aerobactin siderophore | 5’-GGCTGGACATCATGGGAACTGG-3’ | 5’-CGTCGGGAACGGGTAGAATCG-3’ | 300 |
| papC | Pyelonephritis associated pili | 5’-ATATCCTTTCTGCAGGGATGCAATA-3’ | 5’-GTGGCAGTATGAGTAATGACCGTT-3’ | 200 |
| cnf1 | Cytotoxic necrotizing factor 1 | 5’-TCACGGGAATGAACTTATCACCC-3’ | 5’-GTGACATGGCAAAATGATTACAGC-3’ | 498 |
| Antimicrobial resistance | ||||
| blaCTX-M | Antimicrobial resistance | 5’-GACGATGTCACTGGCTGAGCTTAGC-3’ | 5’-AGCCGCCGACGCTAATACA-3’ | 499 |
| blaOXA | 5’-GGCACCAGATTCAACTTTCAAG-3’ | 5’-GACCCCAAGTTTCCTGTAAGTG-3’ | 564 | |
| blaSHV | 5’-GATGAACGCTTTCCCATGATG-3’ | 5’-CGCTGTTATCGCTCATGGTAA-3’ | 214 | |
Primers Used in This Study
3.3. Virulence and Resistance Genes Detection
Virulence genes fimH, traT, cnf1, iutA, and papC were detected by multiplex PCR in a volume of 20 µL with minor modifications (Table 1). PCR conditions for virulence genes were: 4 minutes at 94°C followed by up to 30 cycles of 94°C for 1 minutes, 59°C for 10 seconds, 72°C for 30 seconds, and finally, 72°C for 5 minutes. Furthermore, antibiotics resistance genes (blaCTX-M, blaOXA, and blaSHV) were amplified under the following conditions: 4 minutes at 94°C followed by up to 30 cycles of 94°C for 1 minute, 59°C for 10 seconds, 72°C for 30 seconds, and finally, 72°C for 5 minutes. PCR products were analyzed by electrophoresis in agarose gels (2%) for 1 hour at 60 V/cm and visualized with ethidium bromide and documented by using the Image Lab software. For all agarose gels, the Olerup SSP® DNA Size Marker was used as a molecular length marker (Sweden).
3.4. Statistical Analyses
χ2 and Fisher tests were carried out to compare the populations of ambulatory and hospitalized patients positive to E. coli in urine culture and their relationship with other variables (gender and origin). For statistical analysis of distribution related to the virulence and antimicrobial resistance genes, the following issues were considered: Null hypothesis (H0) = independent variable (presence of detected genes in strains), alternative hypothesis (H1) = dependent variable (detected genes related to the strain or type of gene). Considering P > 0.05, the null hypothesis is accepted (significant) and the alternative hypothesis is rejected. As in all cases, the results were greater than 0.05; therefore, it was concluded that the results were significantly different at 95% confidence, indicating that there was no relationship between strains and the presence of detected genes.
4. Results
4.1. Description of the Study Population
A total of 107 Mexican patients with urinary tract infection by E. coli were treated from April 2016 to January 2017. The patients attending at HJM were included after obtaining their written informed consent. The financial support for the research was provided by the HJM. All patients were classified by gender and origin (ambulatory and hospitalized). The majority of the patients (71/66.35%) were female patients in comparison to 36 (33.64%) male patients. Age ranges of the study population were 24 to 56 and 23 to 34 years old for male and female, respectively. The proportions for ambulatory and hospitalized patients per gender were: 52.11% (n = 37) versus 47.88% (n = 34), and 52.77% (n = 19) versus 47.22% (n = 17) in female and male patients, respectively. Statistical analysis showed that no significant differences were found between hospitalized and ambulatory patient groups (P > 0.05).
4.2. Antimicrobial Resistance Assay
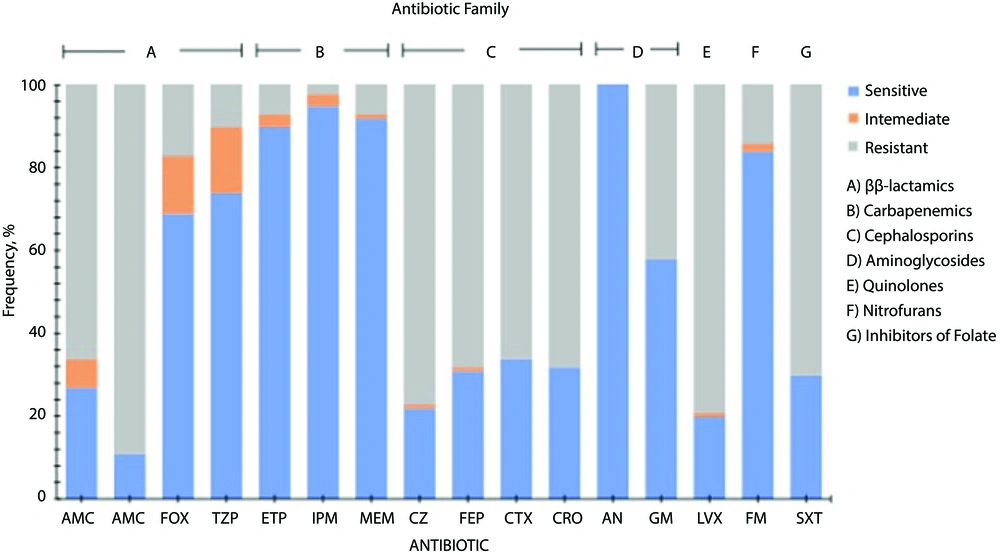
Escherichia coli strains showed differences in susceptibility and resistance patterns to the antimicrobials tested. These results showed that some β-lactams (cefoxitin, piperacillin/tazobactam) carbapenems, aminoglycosides, and nitrofurans were the antimicrobials with the best antimicrobial activity against all strains tested. Penicillins such as amoxicillin/clavulanic acid, and ampicillin, cephalosporins, quinolones, and inhibitors of folate showed lower inhibitory activity on the tested strains. Resistance patterns for all E. coli strains are shown in Figure 1.
Antimicrobial resistance profile of Escherichia coli strains isolated of mexican patients with urinary infection. Antibiotics tested: Amoxicillin/clavulanic acid (AMC), ampicillin (AM), cefoxitin (FOX), piperacillin/tazobactam (TZP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), cefazolin (CZ), cefepime (FEP), cefotaxime (CTX), ceftriaxone (CRO), amikacin (AN), gentamicin (GM), levofloxacin (LVX), nitrofurantoin (FM), trimethoprim/sulfamethoxazole (SXT).
4.3. Bacterial Phylogenetic Assignment
The phylogenetic group B2 (42.05%) was the most predominant, followed by group A (27.1%), group D (24.29%), and finally, group B1 (6.54%). No statistically significant differences were observed between the phylogenetic groups identified by the type of the patient (ambulatory and hospitalized). The distribution of the phylogenetic groups of E. coli strains is shown in Table 2.
| Phylogenetic Group | Strains Origin | Total | |
|---|---|---|---|
| Hospitalized | Ambulatory | ||
| A | 11 (37.93) | 18 (62.06) | 29 (27.10) |
| B1 | 4 (57.14) | 3 (42.85) | 7 (6.54) |
| B2 | 23 (51.11) | 22 (48.88) | 45 (42.05) |
| D | 13 (50.00) | 13 (50.00) | 26 (24.29) |
| Total | 51 (47.66) | 56 (52.33) | 107 (100) |
Phylogenetic Group’s Distribution of Escherichia coli Strains Isolated of Hospitalized and Ambulatory Mexican Patients with Urinary Infectiona
4.4. Virulence Genes Distribution by Phylogenetic Group and Origin
All strains were subjected to a multiplex amplification by PCR in order to detect virulence and antimicrobial resistance genes. The frequencies of virulence genes identified in the E. coli strains were: fimH (92/85.98%), iutA (68/63.55%), traT (66/61.68%), papC (36/33.64%), and cnf1 (15/14.02%) (Figure 2A and B). Eight strains (7.47%) carrying all virulence genes were identified; these strains belonged to the phylogenetic groups B2 (n = 7) and D (n = 1) (Table 3). The proportion of the detection genes was calculated as the number of positive E. coli strains between total strains from the phylogenetic group. The proportion of the identified virulence genes was 1.7 and 1.4 for phylogenetic groups A and B1, respectively and no statistically significant difference was found in the virulence gene frequency from strains that belonged to these phylogenetic groups (P > 0.5).
| Phylogenetic Group E. coli/ Origin | Strains, No. | Virulence Genes | Resistance Genes | ||||||
|---|---|---|---|---|---|---|---|---|---|
| fimH | iutA | traT | papC | cnf1 | blaCTX-M | blaOXA | blaSHV | ||
| A | |||||||||
| Ambulatory | 18 | 16 (88.9)b | 7 (38.9) | 10 (55.6) | 0 (0.00) | 0 (0.00) | 10 (55.6)b | 5 (27.8)b | 1 (5.6)b |
| Hospitalized | 11 | 5 (45.5) | 4 (36.4) | 8 (72.7) | 0 (0.00) | 0 (0.00) | 5 (45.5) | 2 (18.2) | 0 (0.00) |
| B1 | |||||||||
| Ambulatory | 3 | 2 (66.7) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Hospitalized | 4 | 3 (75.0) | 2 (50.0)b | 2 (50.0)b | 1 (25.0)b | 0 (0.00) | 2 (50.0)b | 1 (25.0)b | 0 (0.00) |
| B2 | |||||||||
| Ambulatory | 22 | 21 (95.5) | 18 (81.8) | 8 (36.4) | 13 (59.1) | 7 (31.8) | 9 (40.9) | 9 (40.9) | 0 (0.00) |
| Hospitalized | 23 | 22 (95.7) | 22 (95.7) | 20 (87.0)b | 17 (73.9) | 7 (30.4) | 22 (8.7)b | 19 (82.6)b | 1 (4.3)b |
| D | |||||||||
| Ambulatory | 13 | 12 (92.3) | 5 (38.5) | 9 (69.2) | 3 (23.1) | 0 (0.00) | 5 (38.5) | 6 (46.2) | 0 (0.00) |
| Hospitalized | 13 | 11 (84.6) | 10 (76.9)b | 9 (69.2) | 2 (15.4) | 1 (7.7)b | 8 (61.5) | 7 (53.8) | 0 (0.00) |
Virulence and Antimicrobial Resistance Genes Distribution by Phylogenetic Groups and Origin (Hospitalized and Ambulatory) in Escherichia coli Strains Isolated of Mexican Patients with Urinary Infectiona
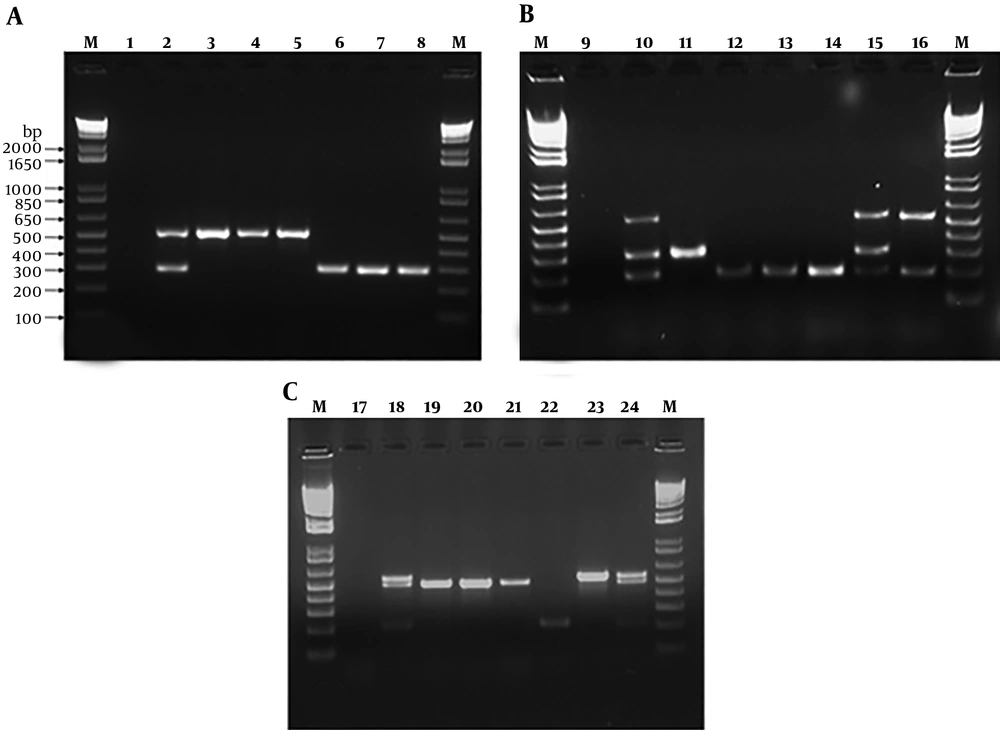
A, electrophoresis of virulence genes (fimH (508 bp) and traT (290 bp)) by duplex PCR of Escherichia coli strains. Lane M: 1 Kb DNA marker; lane 1: Negative control; lane 2: Positive control; lane 3 - 8: 30, 40, 87, 64, 125 and 192 E. coli strains. B, electrophoresis of virulence genes (iutA (300 bp), papC (200), cnf1 (498 bp)) by triplex PCR of E. coli strains. Lane M: 1 Kb DNA marker; lane 9: Negative control; lane 10: Positive control; lane 11 - 16: 17, 32, 96, 98, 95, 52 E. coli strains. C, electrophoresis of antibiotic resistance genes (blaCTX-M(499 bp), blaOXA (564 bp), and blaSHV (214 bp)) by triplex PCR of E. coli strains. Lane M: 1 Kb DNA marker; lane 17: Negative control; lane 18: Positive control; lane 19 - 24: 40, 84, 114, 200, 16, 132 E. coli strains
In contrast, the highest frequency rates were found in the phylogenetic groups B2 and D; proportions were 3.5 and 2.4, respectively. Representing two-fold and 2.5 higher than A (P < 0.05) and B1 (P < 0.001) phylogenetic groups (for group B2). The proportion of virulence genes from the phylogenetic group D was 1.7-fold higher than group B1 (P < 0.05). Significant differences were identified (P < 0.05) in the majority of genes detected in hospitalized patients; iutA, traT, and papC in group B1, traT in group B2, and fimH and cnf1 in group D. Finally, virulence factor fimH presented significant differences in the ambulatory patients in group A.
4.5. Antimicrobial Resistance Genes Distribution by Phylogenetic Groups
The frequencies of the virulence genes identified in E. coli strains were: blaCTX-M(41/38.32%), blaOXA (49/45.79%), and blaSHV (2/1.87%) is shown in Figure 2C. One strain (0.93%) carrying all antimicrobial resistance genes was identified; this strain belonged to the phylogenetic group B2. A summary of antimicrobial resistance genes distribution by phylogenetic groups detected in all strains is shown in Table 3. Proportion of antimicrobial resistance genes was 0.8 and 0.4 for phylogenetic groups A and B1, respectively. The frequencies of antimicrobial resistance genes were 1.3 and 1.0 in the phylogenetic groups B2 and D, respectively. The frequency of antimicrobial resistance in the phylogenetic group B2 was 1.3, representing 1.6-fold and 3.2-fold higher than the phylogenetic groups A (0.8) and B1 (0.4), respectively. Significant differences were identified (P <0.05) in the majority of genes detected in ambulatory patients; blaCTX-M, blaOXA, and blaSHV in group A and B2 and blaCTX-Mand blaOXA in group B1 of hospitalized patients.
4.6. The Relationship Between Virulence and Antimicrobial Resistance Genes in E. coli Strains
The relationship between virulence and antimicrobial resistance genes was evaluated in all E. coli strains. The genetic associations: fimH/blaCTX, fimH/blaOXA, traT/blaCTX, traT/blaOXA, iutA/blaCTX, and iutA/blaOXA showed higher incidence, while the associations with genes cnf1 and papC showed lower incidence in the strains evaluated. Finally, there was no association of the virulence gene cnf1 with the blaSHV resistance gene. The relationship between virulence and antimicrobial resistance genes in E. coli strains is shown in Table 4.
The Relationship Between Virulence and Antimicrobial Resistance Genes in Escherichia coli Strains Isolated of Mexican Patients with Urinary Infectiona
5. Discussion
Urinary tract infections are mainly caused by Gram-negative and facultative anaerobic bacilli such as UPEC (31). In this work, we reported the detection of the main phylogenetic groups of E. coli in the patients (ambulatory and hospitalized) with UTI and the presence of virulence and antimicrobial resistance genes. Data in this work agree with previous reports that indicate that female gender is the most susceptible population to develop UTI. The highest frequency is attributed to this gender due to the anatomy of the vaginal and anorectal region. It has been reported that 80% of the UTIs is caused by UPEC that are different from CEC strains that normally inhabit the gastrointestinal tract (32, 33). Phylogenetic analysis of E. coli strains was divided into four mains groups (A, B1 B2, and D) (34, 35).
The results suggest that most of UTI in Mexican population were mainly caused by E. coli of the phylogenetic group B2. Furthermore, ambulatory and hospitalized patients were infected by E. coli phylogenetic groups A, B1, B2, or D with the same frequency (regardless of the origin of the patient). Virulent and resistant strains are mainly included in the phylogenetic groups B2 and D, whereas commensal ones belong to group A. In previous reports, it was indicated that virulent E. coli strains belonged to B2 and D groups, whereas isolates that belonged to group A were CEC strains and could acquire genes of virulence and antimicrobial resistance by horizontal gene transfer; therefore, they can cause infection (36). Interestingly, we found virulence genes (fimH, iutA, and traT) in CEC (A and B1 groups); the high frequency of fimH in the isolates confirms the importance of the participation of this gene in the initial stage of the infection, since this protein recognizes receptors of the bladder cells; a previous report indicated that mannose-binding type 1 pili, codified in fimH gene, is an important virulence factor involved in the establishment of UTI (37).
On the contrary, the IutA protein is the most important receptor during the infection that contributes to bacterial colonization; this gene had a high frequency detected in pathogenic strains isolated from hospitalized patients. The same phenomenon was observed for the traT gene. The papC and cnf1 genes encoded for a P-fimbriae and cytotoxic necrotizing factor type 1, respectively; these genes were detected in low frequencies in strains belonging to the phylogenetic group A. The absence of virulence genes in CEC is a typical group characteristic; however, the study of the strains that presented these genes (papC gene) is interesting to elucidate the reason why they have these genes to be non-pathogenic strains. The importance of the study of the papC gene in pathogenic and non-pathogenic isolates acquires relevance since this gene is associated with adherence; therefore, its detection provides necessary information about the capacity of these strains to cause infections in the high urinary tract (38).
The simultaneous detection of the papC genes, cnf1 in 31 strains, indicate the possible presence of the pathogenicity island IIJ96 (39), since the presence of these genes is strongly associated with strains that produce UTI cases. However, it is necessary to perform the detection of the hlyA gene, which encodes a hemolysin. The frequency of detection of this genetic binomial in our work was high, compared to other works previously reported (40-42). Virulence and resistance genes were more frequent in strains isolated from hospitalized patients compared to ambulatory patients. We hypothesized that in hospitalized patients, the use of antibiotics in the nosocomial environment might allow bacteria to resist the transfer of genetic material between bacteria by conjugation, transformation or transduction.
Recent work indicates that urine is a rich source of bacteriophages that could participate in the transfer process of DNA (43). Due to the detection of antimicrobial resistance genes in CEC strains, we can speculate that these strains could have participated as receptors in processes of genetic material transfer containing resistance or virulence genes. Since E. coli has a broad phylogenetic diversity, it has a high degree of genome plasticity, with gene losses and gains, through horizontal transfer of DNA (44). Therefore, the gain of genes might allow the commensal bacteria to cause infection due to the acquisition of new genetic material. The differences identified in the phenotype of resistance to antibiotics in the tested strains revealed that the antibiotics of the aminoglycoside and carbapenems families could be the drugs of choice for the treatment of UTI in the population studied, since the strains tested showed greater sensitivity.
The beta-lactam antibiotics, cephalosporins, nitrofurantoins, and inhibitors of folate metabolism, showed little activity against the strains tested. Correlation between the detection of the bla genes and the phenotype against the antibiotics tested was observed. The analysis of the relationship between virulence and resistance genes showed that the blaCTX and blaOXA genes were strongly associated with the presence of the fimH, traT, and iutA genes. The virulence/resistance relationship with the blaSHV gene could not be detected due to the low frequency of this gene in all the isolates. It is clear that the increased virulence and the emergence of antibiotic resistance often arise simultaneously; however, its genetic connection has been little studied, thus it is interesting to know the existence of a possible synergism between these two characteristics in pathogens of clinical interest. The results presented in the current work demonstrated the wide diversity in the distribution of virulence and antibiotic resistance genes between the UPEC and CEC strains.
5.1. Conclusions
In conclusion, the phylogenetic group, the virulence factors, and the susceptibility to antibiotics of E. coli causing UTI infections varied significantly among the Mexican populations, In this regard, the predominant phylogenetic groups causing UTI with virulence and resistance properties were the B2 and D.