1. Background
Tuberculosis is a bacterial disease caused by Mycobacterium tuberculosis, which often affects the lungs but can affect other parts of the body (1). It is one of the biggest health challenges all over the world, particularly in developing countries, which have large unregulated private sectors and weak health systems (2). The number of 10.4 million new cases was estimated according to the WHO report in 2016 around the world, 90% of which were in adults, 65% in males, 35% in females, and 56% in five countries including India, Indonesia, China, Philippines, and Pakistan (3). Tabriz is located in the northwest of Iran neighboring with the Republic of Azerbaijan that is one of the countries with a high rate of multidrug-resistant tuberculosis (4). The transmission route of tuberculosis is from person to person through the air. The identification of the transmission routes and sources of M. tuberculosis is associated with prohibiting the generation of infectious particles, prevention programs, controlling the disease, and reducing or eliminating tuberculosis (5).
The low rate of tuberculosis transmission among the population is the characteristic of most low-incidence countries, but cross-border immigration from high-incidence countries can affect the incidence and features of tuberculosis in the low-incidence countries (6). The increased migration of high-burden tuberculosis countries such as Afghanistan and the Republic of Azerbaijan has resulted in the increased frequency and resistance of tuberculosis in low-burden cities of Iran as a low-incidence country (7, 8). Genotyping approaches of M. tuberculosis are used for epidemiological study, which include DNA fingerprinting methods such as Insertion Sequence 6110-restriction fragment length polymorphism (IS6110-RFLP), variable number of tandem repeats (VNTRs) based on mycobacterial interspersed repetitive units (MIRU-VNTR), exact tandem repeat loci-VNTR (ETR-VNTR), whole genome sequencing (WGS), and single nucleotide polymorphisms (SNPs) (9-13). These genotyping approaches are widely used in the control programs of tuberculosis to determine possible epidemiological links between patients, virulence patterns, and dynamics of transmission and to distinguish between relapse and re-infection (13).
Among the various genotyping approaches, IS6110-RFLP is the “Gold standard” approach, because it has high discriminatory power. However, IS6110-RFLP is a less reliable indicator of clonality for low-copy-number strains, particularly those with fewer than six copies of IS6110, and its profiles are difficult to compare across the results of laboratories (14, 15). The MIRU-VNTR approach investigates the number of repeated mycobacterial interspersed repetitive units based on polymerase chain reaction (PCR) at 12, 15, and 24 loci (11, 16). Mycobacterial interspersed repetitive units are composed of 40 - 100 bp scattered repetitive sequences over 41 locations of entire the chromosome of M. tuberculosis (11). The discriminating power of the MIRU-VNTR typing approach is generally associated with the increasing number and sets of loci used (17, 18). Moreover, the MIRU-VNTR approach is useful for studying at the relatively high resolution of clonal expansion and diversity of particular strains or lineages (19, 20). In addition, ETRs are three or five tandem repeats that are combined by 12 and 24 loci MIRU-VNTR for genotype family classification of M. tuberculosis strains and have less discriminatory power than IS6110 RFLP (21, 22).
2. Objectives
In the present study, we aimed to compare the association of two groups of isolates collected from tuberculosis patients referring to the East-Azarbayjan Center for Tuberculosis in 2006 and 2016 by MIRU-VNTR and ETR-VNTR typing methods. Moreover, we assessed the historical connection among strains and predicted future outbreaks or cluster investigation.
3. Methods
From February 2005 to February 2006, 166 M. tuberculosis isolates were collected from the East Azarbaijan Center for Tuberculosis. From 2015 to 2016, 119 M. tuberculosis isolates were collected from patients with tuberculosis in the East Azarbaijan Tuberculosis Center. Demographic data of patients were collected by questionnaires filled out by patient interviews or based on their documents. All samples were approved as M. tuberculosis isolates by conventional biochemical tests based on catalase, niacin, nitrate reduction, and pigment production of isolates. All isolates were analyzed for antibiotic sensitivity patterns by the proportional method using isoniazid, streptomycin, rifampin, and ethambutol (Hortel, Spain) (23).
We conducted DNA extraction by the Phenol-Chloroform method as previously described (24). All the extracted DNAs were stored at -70ºC for further analysis. For molecular analysis, we used the 12-loci MIRU-VNTR method plus ETR-VNTR (22, 25). The Supply et al. protocol was used for MIRU-VNTR typing of the isolates (26). In this regard, 12 selected loci were amplified by the primer-specific PCR method. All reactions were done in 20 µL volumes using the Takara master mix (Takara, Japan). The thermal protocol included 94ºC for 7 min, followed by 35 cycles of 94ºC for 45 s, and different temperatures for annealing at 45ºC and 72ºC for 55 s. The annealing temperatures were as follows: loci 2 (65ºC), 4 (63ºC), 10 (68ºC), 16 (65ºC), 20 (59ºC), 23 (67ºC), 24 (59ºC), 26 (65ºC), 27 (64ºC), 31 (63ºC), 39 (68ºC), 40 (65ºC), ETR-A (66ºC), ETR-B (68ºC), and ETR-C (69ºC). Electrophoresis of the PCR products was done on the 1.5% agarose gel, stained with ethidium bromide (Merck, Germany), and visualized by UV light in a Manual Gel Documentation System (inGenius3, UK). All sizes of fragments were determined by comparing them with a 50-bp DNA ladder size marker (Yekta Tajhiz, Iran) (26). Mycobacterium tuberculosis PT71 was provided by the Tabriz reference tuberculosis lab as the reference strain and used as external quality control for consistency of the results of MIRU and ETR.
3.1. Statistical Analysis
The phylogenic analysis of the isolates was done using Multi-variable Statistical Package (MVSP) software and MIRU-VNTRplus free web-based software (available at https://www.miru-vntrplus.org/MIRU/index.faces). The Neighbor-joining (NJ) algorithm was used for all comparisons. The categorical distance was used for the analysis of our data (27). A cluster in this study was defined as two or more isolates with identical MIRU-VNTR and ETR patterns while isolates with different patterns in any loci were considered non-clustered isolates. Clusters were assumed to be the result of recent transmission. The chi-square test was used for statistical analysis and p values of < 0.05 were considered significant. The minimum estimation of recent transmission of tuberculosis was calculated by the following formula: (number of clustered isolates-number of clusters)/number of all isolates.
4. Results
4.1. Sample Collection in 2006
In 2006, 197 isolates of M. tuberculosis were collected. Thirty-one isolates had not acceptable DNA for molecular analysis; thus, 166 isolates were analyzed by the MIRU-VNTR method. Ninety isolates (54.27%) were from males and 76 (45.78%) from females and the age of the patients ranged from 2 to 88 years. There was no significant difference in risk factors between clustered and non-clustered patients (P > 0.05). In the MIRU-ETR analysis of these isolates, 104 different patterns were observed including 75 unique patterns and 29 shared patterns with several isolates; 91 isolates were clustered in 29 different clusters. There were 29 isolates from Urmia and 61 isolates from Tabriz in the clusters while no patient isolate from Republic of Azerbaijan was in the clusters. We observed 17 clusters with two members, three clusters with three members, three clusters with four members, one cluster with five members, four clusters with six members, and one cluster with seven members. The biggest cluster included six isolates from Tabriz and one from Urmia.
4.2. Sample Collection in 2016
In 2016, 119 isolates of M. tuberculosis were isolated. All the isolates had enough DNA for further molecular typing. Fifty-eight (48.73%) isolates were from males and 61 (51.26%) from females and the age of patients ranged from 15 to 86 years. Risk factors had no difference between clustered and non-clustered patients (P > 0.05). Twenty-nine isolates were from Republic of Azerbaijan. Eight isolates from Azerbaijan were resistant to at least two antibiotics (five isolates to three antibiotics and three isolates to two antibiotics) and only one isolate from Tabriz was resistant to at least two antibiotics. Only these isolates had resistance to antibiotics. No resistance to streptomycin was observed (Appendix 2 in Supplementary File). Ninety-eight different patterns were observed with 86 unique patterns and 12 shared patterns clustered in 12 different clusters including 33 isolates. Nine clusters had two members, one cluster had four members, one cluster had five members, and one cluster had six members. Patients from Republic of Azerbaijan were included in four clusters that indicated the transmission of tuberculosis among these patients. The biggest cluster included one patient from Republic of Azerbaijan and five patients from Tabriz. Other clusters with patients from Azerbaijan were a cluster with three patients from Republic of Azerbaijan and one from Tabriz and a cluster with one patient from Republic of Azerbaijan and one from Tabriz. In addition, a cluster with only two patients from Republic of Azerbaijan was observed.
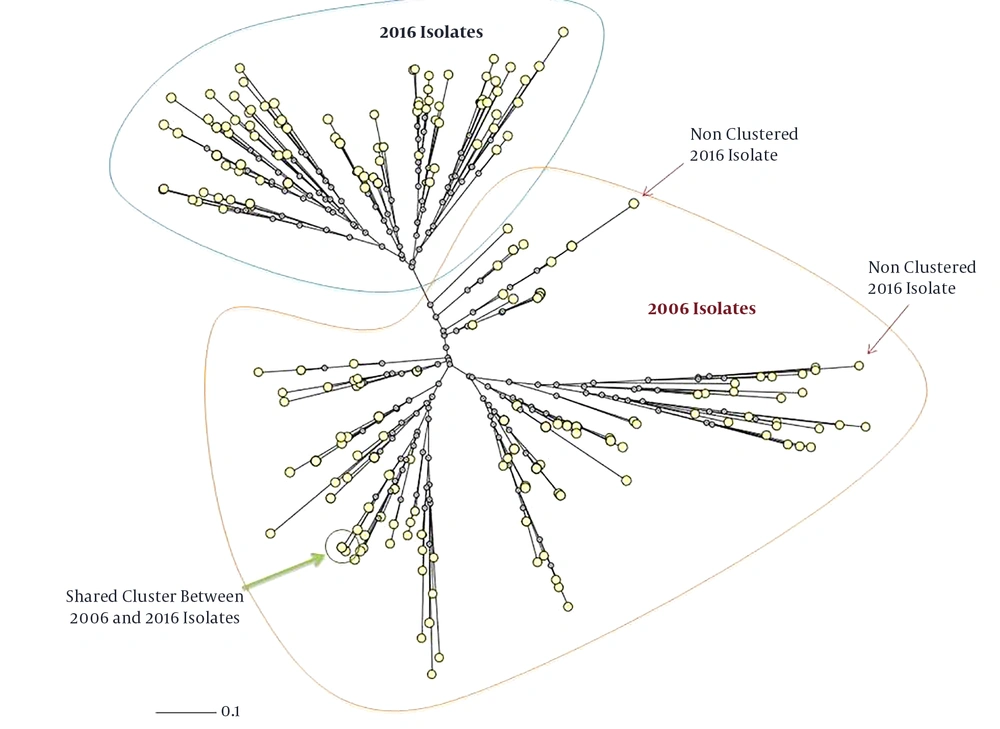
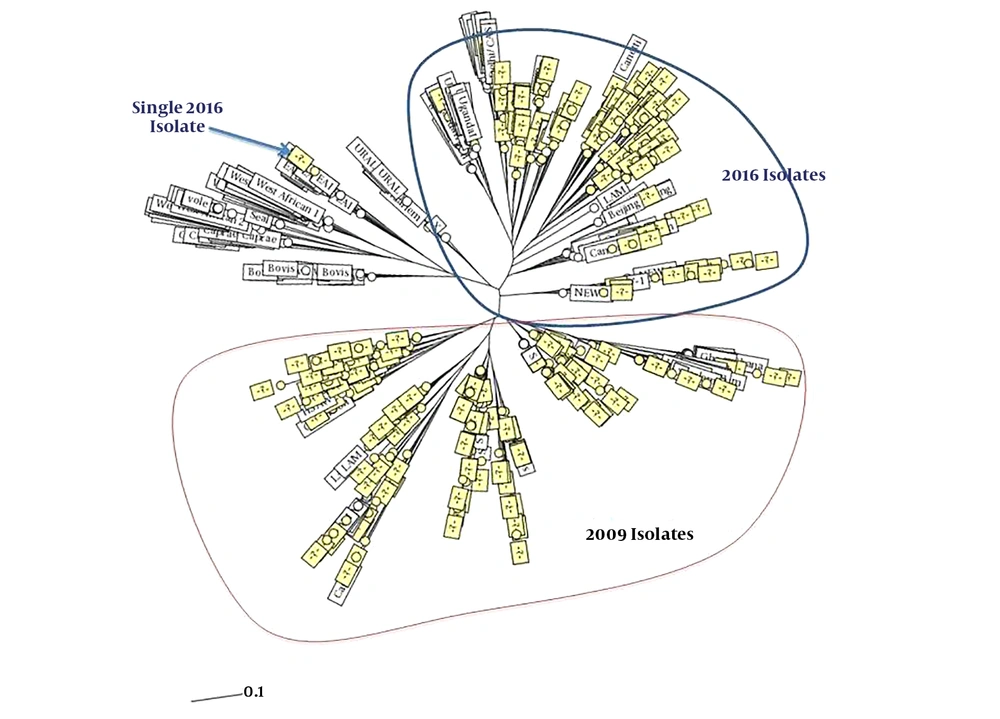
In addition, only three isolates from the 2016 isolates had a close relation and there was a cluster with shared patients from the 2006 and 2016 isolates. It included a patient from Tabriz from the 2016 isolates and a patient from Urmia from the 2006 isolates (Figures 1 and 2). The minimum spanning tree of the isolates showed a close relation among clustered and non-clustered isolates of each sample group from 2006 and 2016 (Figure 3). The comparison among patterns of the present study and other identified international patterns showed that the 2016 isolates had a closer relationship with the international pattern than the 2006 isolates. In the 2006 isolates, the patterns were close to “S” and “LAM” strains and in the 2016 isolates, they were close to “Beijing”, “New Delhi”, and “Uganda” strains (Figure 4). The minimum estimate for the tuberculosis proportion, which is due to the recent transmission of tuberculosis, was 36.7% for the 2006 isolates and 17.6% for the 2016 isolates.
Phylogenic comparison of isolates indicating complete changes in the molecular pattern of isolates during 10 years (neighbor-joining tree provided by MIRU-VNTRplus). Only three isolates from the 2016 isolates had a close relationship with the 2006 isolates, which one of them was in a shared cluster with one from the 2006 isolates.
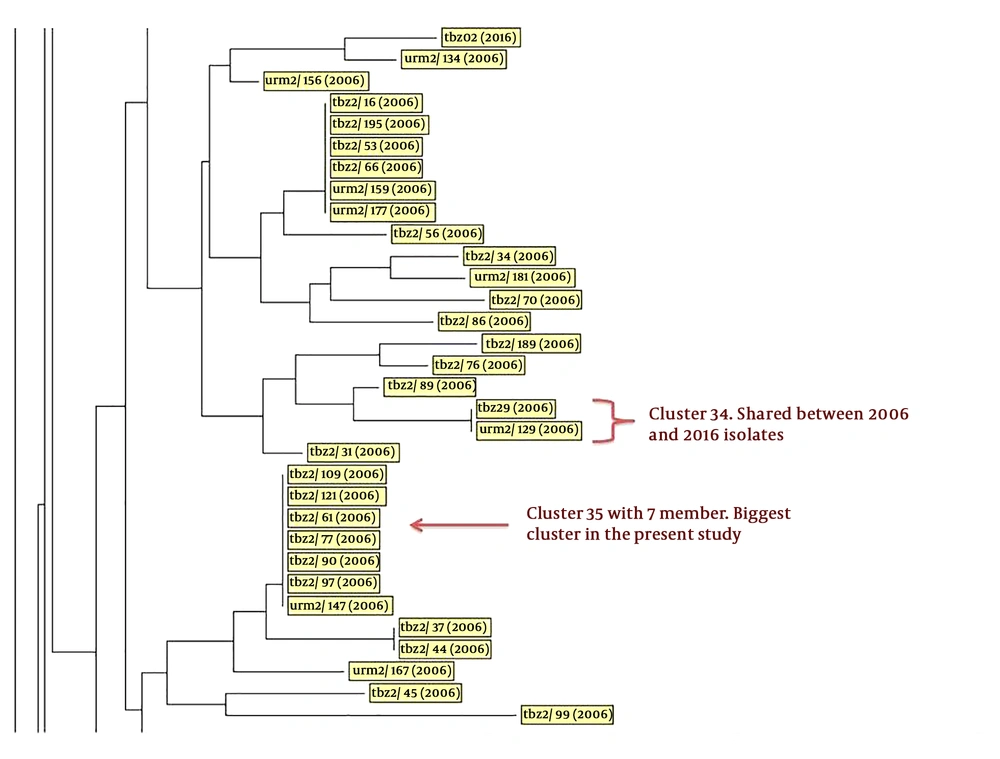
Part of the neighbor-joining phylogenetic tree of isolates. This part reveals a cluster with shared isolates from the years 2006 and 2016. One of these isolates was from Tabriz city and another one from Urmia, which indicates the transmission of isolates between these two patients during this time. Also, a cluster with identity 35 had seven members and was the biggest cluster in this study (the complete tree is provided in Appendix 1 in Supplementary File). Abbreviations indicate the origin of isolates; tbz: Tabriz; urm: Urmia.
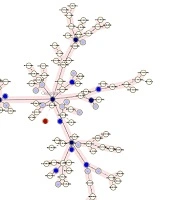
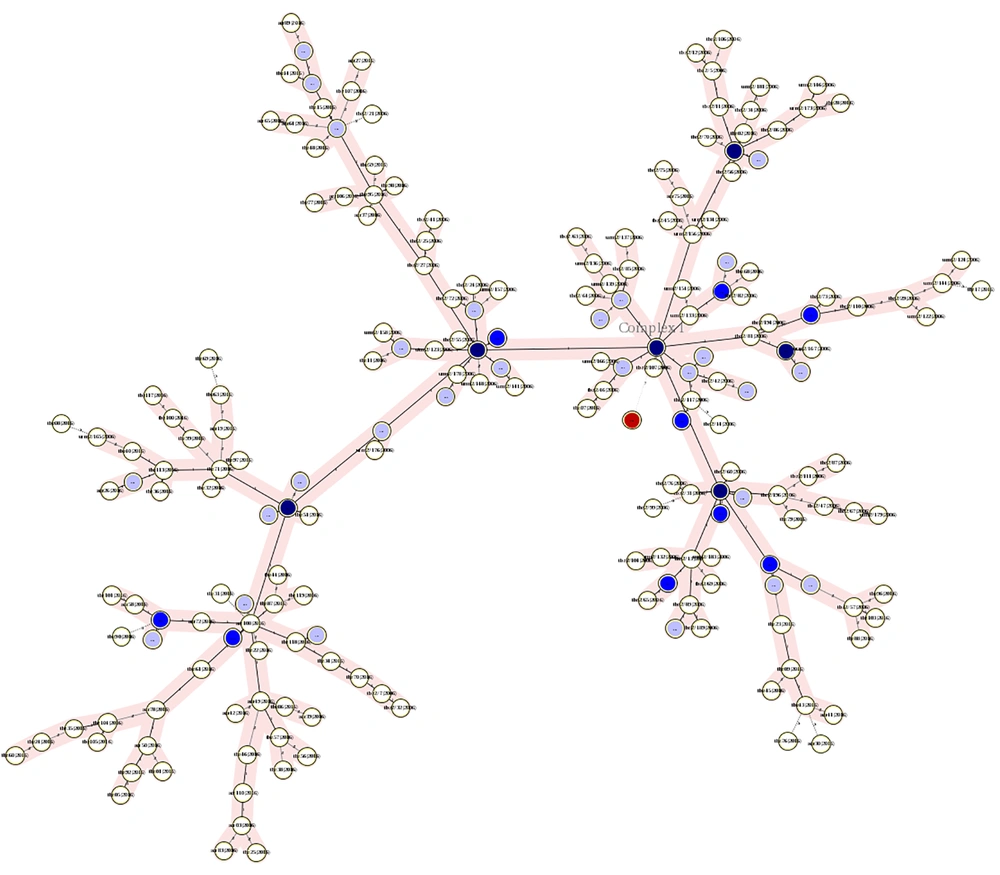
Minimum spanning tree of isolates indicating the number of clusters and their relation with other single isolates. This figure shows how a cluster had a close MIRU-VNTR pattern with non-clustered isolates or other clusters. Red circles are isolates that had no MIRU-VNTR pattern. The darkness of circles indicates the number of isolates in a cluster. Two right branches belong to the 2006 isolates and the left branch belongs to the 2016 isolates.
Comparing patterns with other identified international patterns indicating the closer pattern of the 2016 isolates with the 2006 isolates. The presence of patterns close to the Beijing pattern, which is one of the famous MDR patterns of tuberculosis, indicates that isolates in this region are more the result of the worldwide transmission of tuberculosis, as well as the result of more patients from Republic of Azerbaijan admitted in our region. In the 2006 isolates, the patterns were close to “S” and “LAM” strains and in the 2016 isolates they were close to “Beijing”, “New Delhi”, and “Uganda” strains.
5. Discussion
Phylogenetic analysis plays an important role in the assessment of the evolutionary history of various bacterial strains, such as M. tuberculosis. In addition, the DNA sequences play a key role in the futures of genomic evolution, including adaptation to different host environments, enhanced virulence factors, resistance to antibacterial agents, and other changes in important properties. Repeated sequences are possibly related to the molecular basis of widespread clonal variants and the polymorphism of M. tuberculosis structural genes. We identified 284 M. tuberculosis isolates in both 2006 and 2016 years by using the combination of MIRU-VNTR and ETR-VNTR methods in the northwest of Iran. In the present study, we determined the dynamics of transmission in this region. In the 2006 isolates, we had no Azeri patients referring to the East-Azarbaijan center for tuberculosis, but in the 2016 isolates, we had more admissions of Azeri tuberculosis patients, which caused more MDR isolates. In addition, in 30% of the clusters, we had Azeri patients. Because of the close culture and distance, language similarity, appropriate treatment facilities, and low medical cost, Azeri patients prefer to refer to Tabriz (the capital city of East Azerbaijan province) for the treatment of tuberculosis.
The presence of patterns related to “Beijing”, “S”, and “LAM” strains, which are famous MDR patterns of tuberculosis (28, 29), was directly related to Azeri patients referring to this region. Beijing, S, and LAM strains are more influenced by the worldwide transmission of tuberculosis. Increased cross-border immigration of Azeri patients with more MDR isolates can contribute to the increased risk of tuberculosis among the Tabriz population. The Republic of Azerbaijan is a traditionally prevalent center for Beijing genotypes of tuberculosis (30). However, recent studies indicated an increasing prevalence of Beijing and New Delhi strains in Iran by immigration from Afghanistan to the west and northwest of Iran (31, 32). The increase in the number of genotypes with resistance to antibiotics can cause hardship in infection control that needs attention for control of infection transmission from Republic of Azerbaijan to Iran. These patients prefer treatment in Iran because of free drug and treatment courses in Iran. Providing clinics in borders or inside Republic of Azerbaijan and restricted admission of these patients can control the transmission of infection to the northwest of Iran.
According to the obtained results, three isolates from 2016 had a close relation to the isolates from 2006 and there was a cluster with shared patients from the 2006 and 2016 isolates. These results demonstrated that there were three patients with relapse infection in 2016 who had tuberculosis in 2006. These patients were from different cities in this region; this transmission can indicate the Ice Mountain and show the visible part of the transmission of tuberculosis and the need for tracing possible infected patients untreated and unidentified. Using whole-genome sequencing can help us better understand the changes in the tuberculosis genome during transmission in this time period (33). Figure 2 shows that in cluster 34, we had two patients with an exact similar pattern but from different isolates, indicating the circulation of this isolate in patients and its transmission in this region. The early patient in this cluster had no background of tuberculosis in her family and transmission probably happened in the community.
The minimum estimation was 17% for the 2016 isolates and 36% for the 2006 isolates. The reduced number of isolates in the same period in 2016 and 2006 and less minimum estimate for the tuberculosis proportion indicated less transmission of tuberculosis and the success of tuberculosis transmission control in our region. In our region, the increase in prescribing vitamin D (34), free treatment of tuberculosis, and controlling the treatment trends according to the WHO guidelines (35) have led to the reduction in tuberculosis in 2016 compared to 2006. In an Iranian study from Tehran, it was reported as 24% (36); in other regions worldwide, it was reported from 14.6% (37) in French Guiana to 32% in South African gold miners (38). In a study, the minimum estimation for tuberculosis transmission was 5.1% in patients with negative results on nucleic acid amplification in the United States and it was 11.2% in smear-negative tuberculosis patients (39). These results indicate a reduced rate in our region compared to previous studies.
5.1. Conclusion
The results of the present study indicate tuberculosis controlling in the northwest of Iran was satisfactory but in spite of the reduced number of tuberculosis patients, the isolates were more resistant and had a close relationship with worldwide strains. Cross-border immigration for treatment from Republic of Azerbaijan had significant participation in the recent transmission of tuberculosis in this region. In conclusion, the strict control of patients commuting and developing new tuberculosis clinics inside Republic of Azerbaijan can play a key role in the control of tuberculosis transmission in the northwest of Iran.