1. Background
Hepatitis B virus (HBV) is highly prevalent, as one-third of the world’s population has been exposed to it and 350 - 400 million have developed a chronic infection, with > 1 million deaths per year from cirrhosis and liver cancer (1). In Pakistan, almost more than 9 to 12 million people are estimated to be living with HBV or Hepatitis C virus (HCV), with a carrier rate of 3% - 5% (2, 3). Hepatitis B virus is responsible for more than half of all new cases of liver cancer and it is among the “top ten” causes of cancer death. On the other hand, non-alcoholic fatty liver disease (NAFLD) is currently on a trajectory to become the most common liver disease, as it has affected around 20 - 30% of the global population (4). This renders that the co-occurrence of both diseases is not infrequent and studying this relationship is of pivotal importance.
Hepatitis B has an intimate link with hepatic lipid metabolism. An opposite association is observed between positive Hepatitis B surface antigen (HBsAg) status and the prevalence of fatty liver in humans. It has been suggested that HBsAg seroclearance is more than three-fold higher in those with moderate-to-severe hepatic steatosis than in those without hepatic steatosis (5). A similar observation was reported in mouse models and in vitro (6). Multiple reports have identified variants in patatin‐like phospholipase domain containing 3 (PNPLA3) (I148M) and transmembrane 6 superfamily member 2 (TM6SF2) (E167K) as risk variants for steatosis in patients with NAFLD. However, the data are scarce on the impact of these variants on hepatic steatosis in patients with viral hepatitis, especially in Asian patients with hepatitis B. Notably, it is proposed that these polymorphisms are associated with HBV-DNA levels, suggesting a potential role in HBV persistence (7-9).
In 2015, a single-nucleotide polymorphism (SNP) (rs641738) in membrane-bound O-acyltransferase domain containing 7 (MBOAT7) was identified by a genome-wide association study as a risk variant for alcohol-related cirrhosis (10). The next reports demonstrated that this polymorphism was associated with liver injury in NAFLD, hepatitis C, and hepatitis B (11-13). However, the impact of this variant on steatosis is still less clear.
2. Objectives
The role of PNPLA3 (HGNC:18590), TM6SF2 (HGNC:11861), and MBOAT7 (HGNC:15505) genetic polymorphisms remains obscure in the Pakistani population. Therefore, we aimed to investigate the association of PNPLA3, TM6SF2, and MBOAT7 polymorphisms with hepatic steatosis and hepatitis B persistence in the Pakistani population.
3. Methods
3.1. Patient Cohort
The study comprised 297 Pakistani subjects (240 HBV-infected patients and 57 healthy controls). In the period from July 2016 to May 2017, patients with positive HBsAg tests were recruited. Among 240 chronic HBV patients, 44 were found with steatosis and 196 were non-steatotic. The aspartate aminotransferase platelets ratio index and ultrasound examination were employed to detect and evaluate liver steatosis. The subjects were recruited from the outpatient Department of Hayatabad Medical Complex (HMC), Peshawar, located in the Northwest of the KPK region of Pakistan. The Hayatabad Medical Complex is a major tertiary care hospital in Peshawar that receives a large flow of subjects/patients from all populations of KPK.
3.2. Exclusion Criteria
Patients were excluded if they abused alcohol (≥ 20 g of alcohol daily), had a history of vaccination against HBV and evidence of co-infection with either human immunodeficiency virus (HIV), HCV, hepatitis delta virus (HDV), or other liver diseases.
3.3. Clinical and Laboratory Assessment
The following information was obtained at the time of blood collection: age, gender, ethnicity, alcohol intake, socioeconomic status, vaccination history, and routine laboratory tests. Hepatic steatosis was evaluated based on ultrasound and APRI. Blood samples were collected in EDTA/Heparin vacutainer tubes to avoid clotting. About 4 mL of the whole blood sample was taken from patients visiting the outpatient department and laboratory investigations were carried out at the Department of Microbiology, Quaid-i-Azam University, Islamabad, and HMC, Peshawar. The liver function, cholesterol, triglyceride, and platelets were assessed using Cobas C111 (Roche) following the manufacturer’s instructions and SOPs. The samples were screened for the detection of HBsAg and positive samples were evaluated for anti-HBc-IgM, anti-HBc-IgG, anti-HDV, and anti-HCV. For this purpose, we used commercially available ELISA kits (MBS-SRL, Milano, Italy) according to the manufacturer’s instructions.
3.4. DNA Extraction and Amplification
Viral DNA was extracted and purified using commercially available kits (Sacace Biotechnologies S.R.L, Italy). The genomic DNA was extracted using commercially available DNA extraction kits (GeneJET Thermo scientific) and the Phenol-Chloroform method. A proper protocol was followed for the extraction of human DNA. The extracted human DNA was stored at -80°C. Real-time PCR was done for the viral load using Cepheid Smart cycler.
3.5. Genotyping
Genotyping for PNPLA3 (HGNC:18590) rs738409, MBOAT7 (HGNC:15505) rs641738, and TM6SF2 (HGNC:11861) rs58542926 was undertaken using the TaqMan SNP genotyping allelic discrimination method (Applied Biosystems, Foster city, CA, USA) (14). Genotyping was blinded to clinical variables.
3.6. Statistical Analysis
Data are shown as mean and standard deviation (SD), median, range or number, and proportion, as appropriate. The frequency of PNPLA3 rs738409, MBOAT7 rs641738, and TM6SF2 rs58542926 genotypes was compared between different groups using Fisher’s exact test. The Cochran-Armitage test was used for the assessment of trends. The student’s t-test or non-parametric Wilcoxon-Mann-Whitney U-test or Kruskal-Wallis test was used to compare quantitative data, as appropriate. All tests were two-tailed and P values of < 0.05 were considered significant. Hardy-Weinberg equilibrium tests of PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT rs641738 were performed using the chi-square test. The HBV DNA levels were log-transformed before modeling. The logistic regression model was used and the results are expressed as OR and 95% CI. Statistical analyses were performed using the statistical software package SPSS for Windows, version 21 (SPSS, Chicago, IL).
4. Results
4.1. Patient Characteristics
The demographic, biochemical, and virological characteristics of the studied HBV-infected hepatic steatotic and non-steatotic patients are presented in Table 1. The median age was 29 years and 28 years in HBV patients and controls, respectively. Moreover, 72.4% and 61.4% were males in the patient and control groups, respectively, and 18.3% of the HBV patients had hepatic steatosis.
| Variables | Non-Steatosis (N = 196) | Steatosis (N = 44) | P Value |
|---|---|---|---|
| Ageb, y | 26 (4 - 75) | 44 (17 - 82) | 0.0001c |
| Genderb (male/female) | 142/54 (72.4%) | 27/17 (61.4%) | 0.149 |
| ALTb (IU/L) | 33 (11 - 211) | 175 (25 - 680) | 0.0001c |
| ASTb (IU/L) | 35 (15 - 103) | 149 (70 - 459) | 0.0001c |
| Total bilirubin (mg/dL) | 0.7 (0.3 - 3.8) | 0.9 (0.4 - 6.1) | 0.140 |
| Cholesterol (mg/dL) | 131 (78 - 260) | 138 (78 - 280) | 0.001c |
| TG (mg/dL) | 173 (92 - 351) | 189 (120 - 312) | 0.014c |
| Platelets (× 109/L) | 245 (150 - 415) | 169 (95 - 243) | 0.0001c |
Abbreviations: ALT, alanine aminotransferase; AST: Aspartate Aminotransferase; IU, international unit; TB, total bilirubin; TG: triglyceride.
aContinuous variables were described as median and interquartile range.
bThe relative difference between two groups.
cStatistically significant (P < 0.05).
4.2. PNPLA3 rs738409 and TM6SF2 but not MBOAT rs641738 Were Associated with the Risk of HBV Chronicity
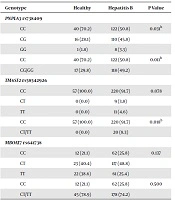
The genotype distribution of PNPLA3 rs738409, TM6SF2, and MBOAT7 in HBV-infected patients and the healthy Pakistani population is presented in Table 2. Genotype distribution was in the Hardy-Weinberg equilibrium in both groups. The minor allele frequency (MAF) (G) of PNPLA3 rs738409 was 0.26, which was significantly higher than that noticed in our healthy cohort (MAF 0.16, P = 0.031); it was also true using the recessive model (P = 0.011). This association remained significant in multiple logistic regression analysis after adjusting for age and gender (OR= 0.41, 95% CI: 0.22 - 0.78, P = 0.006) (Table 3).
| Genotype | Healthy | Hepatitis B | P Value |
|---|---|---|---|
| PNPLA3 rs738409 | |||
| CC | 40 (70.2) | 122 (50.8) | 0.031b |
| CG | 16 (28.1) | 110 (45.8) | |
| GG | 1 (1.8) | 8 (3.3) | |
| CC | 40 (70.2) | 122 (50.8) | 0.011b |
| CG/GG | 17 (29.8) | 118 (49.2) | |
| TM6SF2 rs58542926 | |||
| CC | 57 (100.0) | 220 (91.7) | 0.078 |
| CT | 0 (0.0) | 9 (3.8) | |
| TT | 0 (0.0) | 11 (4.6) | |
| CC | 57 (100.0) | 220 (91.7) | 0.018b |
| CT/TT | 0 (0.0) | 20 (8.3) | |
| MBOAT7 rs641738 | |||
| CC | 12 (21.1) | 62 (25.8) | 0.137 |
| CT | 23 (40.4) | 117 (48.8) | |
| TT | 22 (38.6) | 61 (25.4) | |
| CC | 12 (21.1) | 62 (25.8) | 0.500 |
| CT/TT | 45 (78.9) | 178 (74.2) |
aValues are expressed as No. (%).
bStatistically significant (P < 0.05).
| Genotype | Healthy vs. Chronic Hepatitis B | |
|---|---|---|
| OR (95%CI)a | P Valuea | |
| PNPLA3 rs738409 | ||
| CC | 1 | |
| CG | 0.4 (0.23 - 0.83) | 0.012b |
| GG | 0.3 (0.04 - 3.14) | 0.370 |
| Adjusted | 0.4 (0.22 - 0.78) | 0.006b |
| Dominant | 2.2 ( 1.22 - 4.23) | 0.009b |
| CC | 1 | 1 |
| CG/GG | 0.4 ( 0.23 - 0.81) | 0.009b |
| MBOAT7 rs641738 | ||
| CC | 1 | |
| CT | 1.0 (0.47 - 2.17) | 0.968 |
| TT | 1.8 (0.84 - 4.09) | 0.121 |
| Adjusted | 1.3 (0.66 - 2.70) | 0.417 |
| Dominant | 0.7 (0.38 - 1.54) | 0.454 |
| CC | 1 | |
| CT/TT | 1.3 (0.64 - 2.62) | 0.454 |
aOdds ratio and P values were calculated by using binary logistic regression and multiple logistic regression adjusted for age and gender.
bStatistically significant (P < 0.05).
The MAF of TM6SF2 rs58542926 (T) was 0.06, which was significantly higher than that observed in our healthy cohort (MAF 0.00 (P = 0.078); it was also true using the dominant model (P = 0.018). The T allele was significantly associated with HBV chronicity (OR= 1.09, 95% CI: 1.05 - 1.13, P = 0.018). In contrast, the genotype distribution of MBOAT rs641738 was not significantly different between HBV-infected patients and healthy controls (0.49 vs. 0.41, P = 0.137); this was also true using the recessive model (P = 0.500). This remained the same in multiple logistic regression analysis after accounting for the same variables mentioned above (OR: 1.3, 95% CI: 0.66 - 2.70, P = 0.417) (Table 3).
4.3. PNPLA3 rs738409 and TM6SF2 rs58542926 but not MBOAT rs641738 Were Associated with Hepatic Steatosis
Age, serum cholesterol, and triglycerides were higher in patients with hepatic steatosis though they were insignificant, while significant differences in other clinical variables like ALT, AST, and platelets were also noticed between patients with and without hepatic steatosis, as shown in Table 1. Next, we explored the association of PNPLA3, TM6SF2, and MBOAT7 with hepatic steatosis. The genotype distribution of PNPLA3 rs738409, TM6SF2, and MBOAT7 in chronic HBV-infected patients and Pakistani population with hepatic steatosis is shown in Table 4. The rs738409 GG genotype, observed in 3% of patients, was associated with steatosis (0.27 vs. 0.21, P = 0.033) (OR: 2.79, 95% CI: 0.64 ‐ 12.16; P = 0.063) thought it was insignificant due to the sample size. The rs58542926 TT genotype, observed in 4.6% of patients, the minor allele (T) frequency (MAF) of TM6SF2 rs58542926 was significantly higher in patients with steatosis than in those without steatosis (0.11 vs. 0.05, P = 0.024). The T allele was not significantly associated with steatosis (OR: 1.54, 95% CI: 0.53 - 4.5, P = 0.424). In contrast, the MBOAT rs641738 CC genotype, observed in 25.8% of patients, the genotype distribution of MBOAT rs641738 was not significantly different between chronic HBV-infected patients and steatotic cohort (0.49 vs. 0.47, P = 0.666); it was also true using the recessive model (P = 0.448). This remained the same in multiple logistic regression analysis after adjusting for age and gender (OR: 0.9, 95% CI: 0.37 - 2.49, P = 0.950).
aValues are expressed as No. (%).
bStatistically significant (P < 0.05).
4.4. Association of PNPLA3, TM6SF2, MBOAT7 Genotypes with Clinical Variables
Finally, we examined if baseline clinical variables differed among HBV-infected subjects according to the PNPLA3 rs738409 genotype; the results are depicted in Table 5. We observed that subjects with rs738409 CC genotype were significantly older than those with CG/GG genotype (P = 0.005), while here we did not observe any significant association between the rs738409 genotype (CC versus CT/TT) and any of the clinical variables (i.e., age, gender, ALT, AST, HBV-DNA, total bilirubin, cholesterol, and TG). Similarly, there was no association between TM6SF2 rs58542926 or MBOAT7 rs641738 and any of these clinical variables, except for cholesterol (Table 5).
| Genotypesa | P Valueb | ||
|---|---|---|---|
| PNPLA3 rs738409 | CC | CG/GG | |
| Age | 31.9 ± 14.4 | 26.6 ± 12.1 | 0.005c |
| HBV-DNA log IUd | 5.1 ± 2.1 | 5.4 ± 2.3 | 0.298 |
| ALT (IU/L) | 51.9 ± 60.1 | 52.6 ± 85.4 | 0.935 |
| AST (IU/L) | 53.0 ± 48.4 | 49.4 ± 52.9 | 0.548 |
| Total bilirubin (mg/dL) | 0.79 ± 0.41 | 0.82 ± 0.61 | 0.630 |
| Cholesterol (mg/dL) | 144.6 ± 45.9 | 135.3 ± 42.2 | 0.073 |
| TG (mg/dL) | 177.5 ± 50.6 | 169.4 ± 48.6 | 0.164 |
| Platelets (× 109/L) | 240.6 ± 60.3 | 238.3 ± 62.9 | 0.747 |
| TM6SF2 rs58542926 | CC | CT/TT | |
| Age | 29.2 ± 13.6 | 26.8 ± 13.8 | 0.446 |
| HBV-DNA log IUd | 5.3 ± 2.2 | 4.8 ± 2.0 | 0.432 |
| ALT (IU/L) | 50.2 ± 71.1 | 79.0 ± 88.5 | 0.087 |
| AST (IU/L) | 50.5 ± 49.7 | 63.9 ± 59.9 | 0.252 |
| Total bilirubin (mg/dL) | 0.80 ± 0.51 | 0.84 ± 0.44 | 0.714 |
| Cholesterol (mg/dL) | 140.5 ± 44.2 | 139.2 ± 48.2 | 0.903 |
| TG (mg/dL) | 174.7 ± 50.0 | 165.3 ± 46.8 | 0.431 |
| Platelets (× 109/L) | 240.0 ± 60.9 | 233.7 ± 68.8 | 0.660 |
| MBOAT7 rs641738 | CC | CT/TT | |
| Age | 26.8 ± 13.4 | 29.8 ± 13.5 | 0.100 |
| HBV-DNA log IUd | 5.4 ± 2.2 | 5.2 ± 2.2 | 0.527 |
| ALT (IU/L) | 44.9 ± 48.4 | 54.6 ± 78.9 | 0.323 |
| AST (IU/L) | 43.8 ± 33.9 | 53.9 ± 54.7 | 0.138 |
| Total bilirubin (mg/dl) | 0.82 ± 0.50 | 0.79 ± 0.51 | 0.718 |
| Cholesterol (mg/dL) | 149.4 ± 44.8 | 137.4 ± 43.9 | 0.043c |
| TG (mg/dL) | 180.2 ± 44.0 | 173.8 ± 51.5 | 0.199 |
| Platelets (× 109/L) | 233.1 ± 51.9 | 241.7 ± 64.2 | 0.300 |
aValues are expressed as mean ± SD.
bP values were calculated by using logistic regression.
cStatistically significant (P < 0.05).
dHBV DNA in log 10; IU, international unit.
5. Discussion
In this study, for the first time, we investigated the role of functional polymorphisms in three main lipid-related genes, namely PNPLA3, TM6SF2, and MBOAT7, in hepatic steatosis and HBV chronicity in the Pakistani population. Consistent with other data reported in different populations, we demonstrated that PNPLA3 and TM6SF2 but not MBOAT7 were associated with hepatic steatosis in HBV patients (7, 13, 15). Interestingly, our data suggest that these two polymorphisms may be implicated in HBV persistence. The natural history of viral hepatitis infection including HBV infection exhibited a marked inter-individual variation, indicating a pivotal role for genetic basis in shaping the outcome of such patients (16). The last years have witnessed multiple GWAS studies, which revealed multiple risk variants for common complex diseases including infectious diseases (8, 9, 17-20). However, to date, most studies were conducted in either Caucasian or other Asian populations, with very limited data in the Pakistani population, while Pakistan is a country with a very high prevalence of HBV infection and a large population.
Patients with hepatic steatosis tended to have higher ages and serum cholesterol and triglyceride levels, though not significantly, but no difference was noticed in the HBV-DNA levels. It is consistent with a suggestion that steatosis in HBV is metabolic rather than viral. In this context, a polymorphism in the PNPLA3 gene, which is known as adiponutrin and has triacylglycerol lipase activity, has been shown robustly by multiple studies to have a strong association with liver fat content in NAFLD patients, as well as patients with viral diseases (15, 21, 22). Similarly, nonsynonymous coding variants in transmembrane 6 superfamily member 2 (TM6SF2) (E167K) have recently been well studied as genetic risk factors for hepatic fat accumulation and progression to NASH, fibrosis, and HCC. Similar data were reported with hepatic steatosis in patients with viral hepatitis (7, 8, 14, 23). However, the exact function of TM6SF2 is not known, but it regulates cholesterol synthesis and the secretion of lipoproteins (24). This polymorphism in the TM6SF2 gene (rs58542926) affects hepatic TM6SF2 mRNA and protein expression.
In contrast, the association between polymorphisms in the MBOAT7 and hepatic steatosis is controversial; most of the evidence suggests the lack of an association or a very marginal association at the best. Though the mechanisms of MBOAT7 function are still unclear, it is likely involved in hepatic inflammation via its function in the remodeling pathway of phosphoinositides (Land’s cycle) that assigns arachidonic acids (AAs) to lysophosphatidylinositol; it can also intensify the inflammatory milieu in macrophages and other immune cells (11, 12, 14). Viral hepatitis including HBV has an intimate interaction with lipids (25, 26). Our data suggest that polymorphisms in PNPLA3 and TM6SF2 but not in MBOAT7 might be implicated in HBV persistence. This is consistent with a couple of reports suggesting that these two polymorphisms modulate HBV-DNA (8, 12). Furthermore, the lack of association between MBOAT7 and HBV persistence is consistent with our current data and others that indicate the effect of MBOAT7 on liver diseases is unlikely to be in hepatic steatosis. Further studies are required to understand the functional mechanisms of these effects and if they can be exploited for therapeutic purposes.
5.1. Conclusions
In conclusion, we demonstrated for the first time that polymorphisms in PNPLA3 and TM6SF2, but not in MBOAT7, are associated with steatosis and HBV viral persistence in the Pakistani population. Future direction will be to explore if these findings can aid in guiding efforts for the personalization of medicines and finding novel therapeutic targets.