1. Background
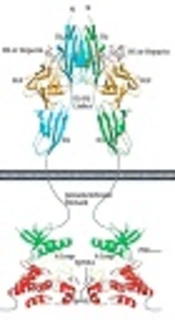
Fibroblast growth factor receptors (FGFRs) belong to the receptor tyrosine kinase (RTK) family and consist of seven distinct FGFRs, i.e., FGFR1b, FGFR1c, FGFR2b, FGFR2c, FGFR3b, FGFR3c and FGFR4. Amongst these, FGFR2b is an epithelial isoform of FGFR2 that regulates key cellular processes, such as cell proliferation, growth, differentiation, migration, and survival (1, 2), playing a crucial role in signal transduction. Human fibroblast growth factor receptor 2b kinase domain has molecular mass of approximately 38 KDa and contains 334 amino acid residues arranged in the N-terminal and the C-terminal domains (Figure 1) (3, 4). Several gain-of-function point mutations of FGFR2b kinase domain have already been identified in different cancers, including breast, ovarian, and uterus (5).
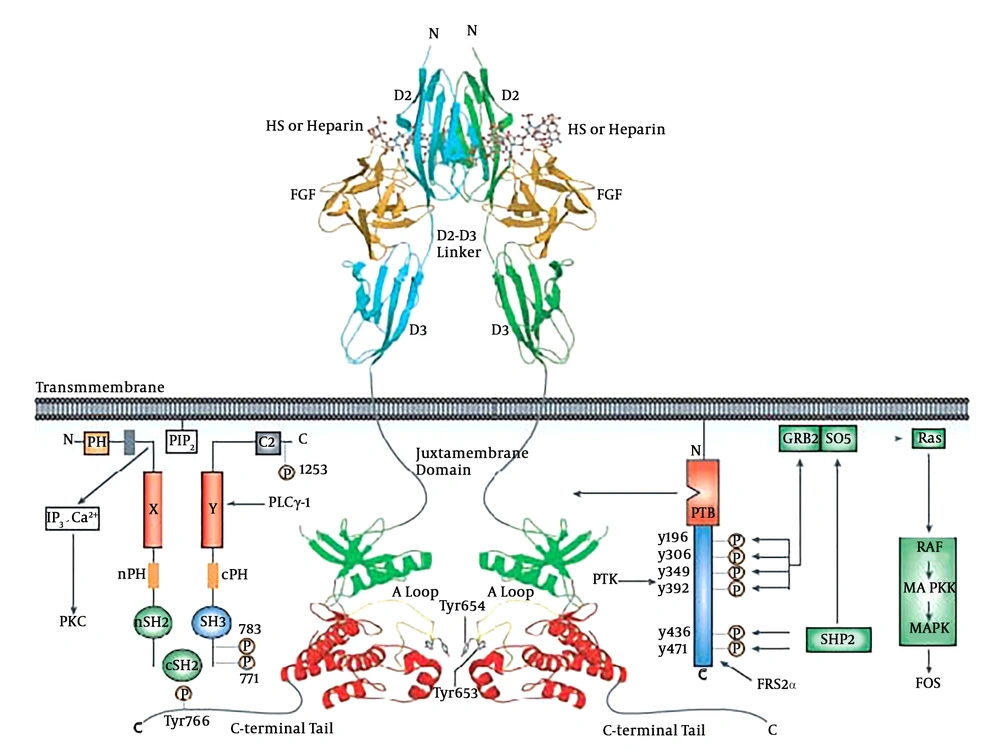
FGF cell signaling. The structural basis by which heparin/HS assists FGF and FGFR to form a symmetric 2:2 FGF-FGFR dimer are shown. The N-terminal and C-terminal lobes of the kinase domain are colored green and red, respectively. Domain organization of PLCγ-1 and FRS2α, two major intracellular targets, are shown (4).
Several epidemiological, in vitro and animal studies (1-3) have previously demonstrated that flavonoids, such as gallic acid, naringenin, quercetin and catechin can affect the growth and proliferation of different human tumor cells, such as breast (6), colorectal (7, 8), and prostate cancer cells (9, 10). These studies, however, have shown contradictory results. Some in vitro studies suggest that flavonoids, gallic acid for example, act as stimulants, enhancing proliferation of cancer cells (11, 12). Other studies, however, have identified some flavonoids, including naringenin, as anti-proliferative agents, showing that they inhibit cell proliferation in certain types of cancer cells (13, 14). Importantly, the mechanism of action, by which flavonoids affect growth and proliferation of cancer cells are not well understood (15, 16). Having said that, some studies have suggested that flavonoids modulate tyrosine kinase activity of different receptor tyrosine kinases, such as insulin-like growth factor receptor (IGFR), epidermal growth factor receptor (EGFR), and platelet-derived growth factor receptor (PDGFR) in different cancer cells (17-19).
2. Objectives
The present study was conducted to obtain better understanding of the effect of flavonoids on FGF cell signaling and also to shed new light on the mechanism of action, by which flavonoids alter the FGF signaling pathway. To this end, this study characterized stability and structural changes of human FGFR2b kinase domain upon interaction with four different flavonoids.
3. Methods
3.1. Chemicals
Gallic acid (GA), naringenin, quercetin and catechin (Sigma-Aldrich Co., Germany) were dissolved in dimethyl sulfoxide (DMSO), prepared from a stock solution stored at 4°C. To induce chemical denaturation, 8 M guanidinium chloride (GdnHCl) solution was prepared. All other reagents used in this study were of analytical grade and purchased from Sigma-Aldrich Co.
3.2. Expression and Purification of FGFR2b Kinase Domain Fragment
The mutated recombinant kinase domain fragment of human FGFR2b was cloned in pLEICS-01 vectors for bacterial expression. The original empty pLEICS-01 vector was provided by the University of Leicester, UK. Additionally, the recombinant vector was previously reconstructed in the researchers’ lab for another project using the ligase independent cloning method (20). The proteins were expressed in E. coli BL21 (DE3) cells with 1 mM Isopropyl Β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich Co., Germany). For purification, the bacterial cells were lysed in 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 8, 10% glycerol, and 100 mM NaCl. The clarified lysate was loaded on a Ni2+-nitrilotriacetic acid (Ni2+-NTA) affinity chromatography (Sigma-Aldrich Co., Germany). The protein was eluted with 50 to 200 mM imidazole gradient. The fractions containing FGFR2b kinase domain were dialyzed in 25 mM Tris-HCl, 100 mM NaCl, PH 7.5, subdivided to smaller aliquots, and stored at -80°C. Purity of fractions were based on SDS–PAGE analyses. The concentrations of FGFR2b kinase domain were determined using Nano Drop spectrophotometry and an extinction coefficient at 280 nm of 41160 M-1 cm-1 (15, 21).
Moreover, in this study, the researchers engineered the kinase domain fragment of human FGFR2b and introduced two mutations in the kinase domain in order to express the protein in its active state. Mutated positions were created in E565A and K660E residues and were selected based on associated pathologic disorders, namely Pfeiffer syndrome and endometrial cancer, respectively (4, 22). To confirm that the protein was in an active sate, interaction of the purified protein with wild type and mutant SH2 domains of phospholipase C (PLC) was studied. The protein sample was dialyzed and then used to analyze its interaction with both the wild type and mutant SH2 domains of PLC, using polyacrylamide gel electrophoresis (PAGE).
3.3. Fluorescence Spectroscopy of FGFR2b Kinase Domain
Intrinsic fluorescence measurements of purified proteins were recorded on a Cary Eclipse spectrofluorometer (Varian, Australia), using a 10-mm path length quartz cuvette. photomultiplier tube (PMT) detector was set to medium voltage. Both excitation and emission slits with a 5-nm band pass were used. The fluorescence spectra were produced with excitation at 280 nm and the emission was monitored in the range of 300 to 345 nm at room temperature. For removing the effect of background intensity of the buffer, the system was zeroed with a blank solution (400 µL of buffer). The protein concentrations (0.15 mg/mL) were kept constant in each sample solution. Flavonoids incubated with protein had constant concentrations of protein and different concentration of gallic acid (25 µM to 500 µM), naringenin (50 µM to 3 mM), quercetin (100 µM to 1 mM) and catechin (100 µM to 500 µM). Furthermore, by adding suitable amounts of buffer containing 25 mM Tris-HCl and 100 mM NaCl, and at PH 7.5, the final volumes of solution were kept 400 µL. Then, the emission spectra were taken after sample incubation for twenty minutes.
3.4. Fluorescence Spectroscopy and Chemical Denaturation of FGFR2b Kinase Domain
In order to induce conformational change in the purified proteins structure, the kinase domain was titrated with guanidine hydrochloride (GdnHCl). To this end, increasing concentration of GdnHCl (0, 0.25, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0, 5.5 and 6 M) was added to the purified protein at room temperature. Each experiment was repeated three times. The excitation wavelength was set at 280 nm.
4. Results
4.1. Expression and Purification of FGFR2b Kinase Domain Fragment
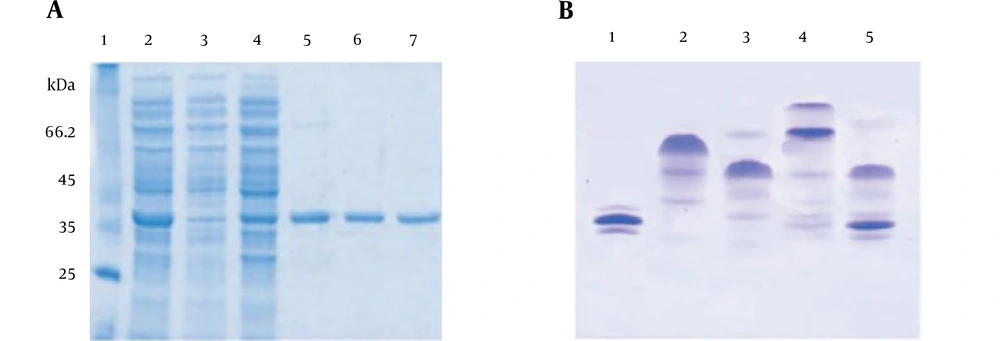
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed that maximum expression of recombinant proteins occurred four hours post induction. The SDS-PAGE analysis in Figure 2A confirmed that no contamination and bacterial proteins were co-eluted with the purified protein after a Ni-NTA affinity chromatography (Figure 2A). In order to prove the purified recombinant protein is active, protein interaction with the SH2 region of phospholipase C gamma (PLC-γ) was investigated by a native PAGE method. As shown in Figure 2B, the native gel analysis clearly demonstrated that the wild type of SH2 domains of PLC-γ (His6-nSH2-cSH2) could interact with the purified tyrosine FGFR2b kinase domain, whereas the mutant SH2 domains of PLC-γ (His6-nSH2 (R562A)-cSH2 (R694A) was not able to bind to the recombinant FGFR2b kinase domain, confirming that the purified protein was in its active state.
A, Purification of the target protein by affinity chromatography. Lane 1 contains high molecular weight markers. Lanes 2 shows samples taken from the load and lanes 3 shows flow through and lanes 4 - 7 show eluted samples. B, Interaction of hFGFR2b with both the wild type and mutant SH2 domains of PLC-γ using native PAGE. Lanes 1, 2, 3, 4 and 5 show the recombinant hFGFR2b kinase domain, His6-nSH2-cSH2, His6-nSH2 (R562A)-cSH2 (R694A), His6-nSH2-cSH2: hFGFR2b kinase domain and His6-nSH2 (R562A)-cSH2 (R694A): hFGFR2b kinase domain, respectively.
4.2. Fluorescence Measurements
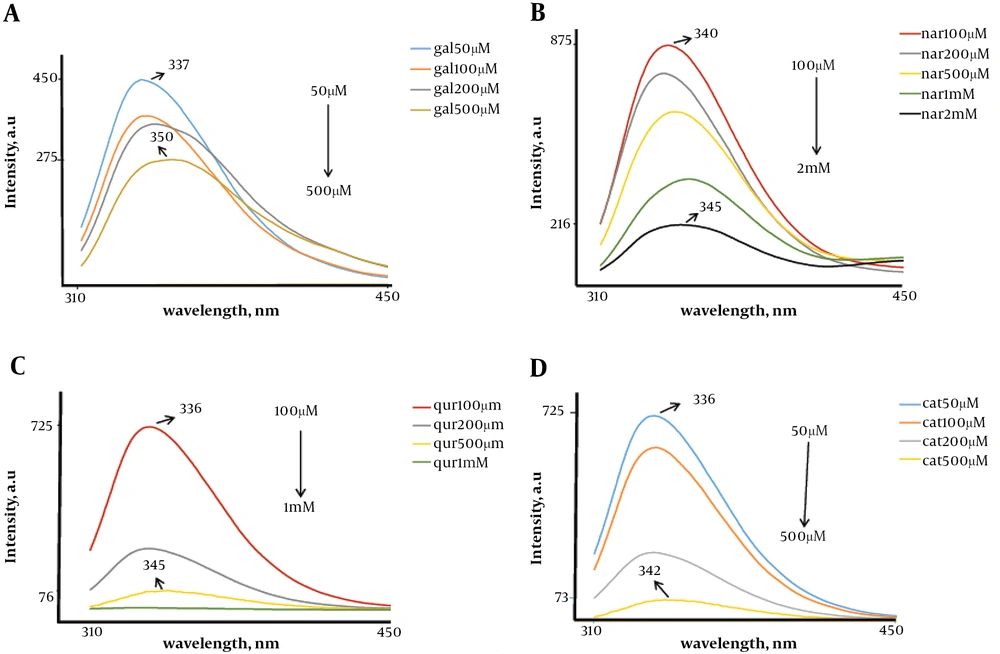
Figure 3A shows the effect of increasing concentrations of gallic acid, from 25 µM to 500 µM, on the fluorescence emission spectrum of native FGFR2b kinase domain fragment. An increase in concentration of gallic acid induced a significant red-shift in the wavelength of maximum emission (from 25 µM to 500 µM) and also increased the fluorescence intensity. Figure 3B shows the effect of increasing concentrations of naringenin, from 50 µM to 3 mM on the fluorescence emission spectrum of native FGFR2b kinase domain fragment. An increase in concentration of naringenin led to a remarkable red-shift in the wavelength of maximum emission (from 50 µM to 3 mM) and also an increase in fluorescence intensity was observed. Figure 3C shows the effect of increasing concentrations of quercetin, from 100 µM to 1 mM, on the fluorescence emission spectrum of native FGFR2b kinase domain fragment. An increase in concentration of quercetin resulted in a considerable red-shift in the wavelength of maximum emission (from 100 µM to 1 mM) and also increase in the fluorescence intensity. Figure 3D shows the effect of increasing concentrations of catechin, from 100 µM to 500 M, on the fluorescence emission spectrum of native FGFR2b kinase domain fragment. As shown in Figure 3D, an increase in concentration of catechin induced a significant red-shift in the wavelength of maximum emission (from 100 µM to 500 µM) and also an increase in fluorescence intensity could be observed.
Fluorescence spectra for FGFR2b kinase domain. A, Represents fluorescence emission spectra at different concentrations of gallic acid (from 50 µM to 500 µM); B, represents fluorescence emission spectra at different concentration of naringenin (from 100 µM to 2 M); C, represents fluorescence emission spectra at different concentration of quercetin (from 100 µM to 1 M); and D, represents fluorescence emission spectra at different concentration of catechin (from 50 µM to 500 µM). Emission shows increase in the intensity of maximum emission spectra with increasing gallic acid concentration.
4.3. Unfolding Assessment of FGFR2b Kinase Domain Undergoing Chemical Denaturation
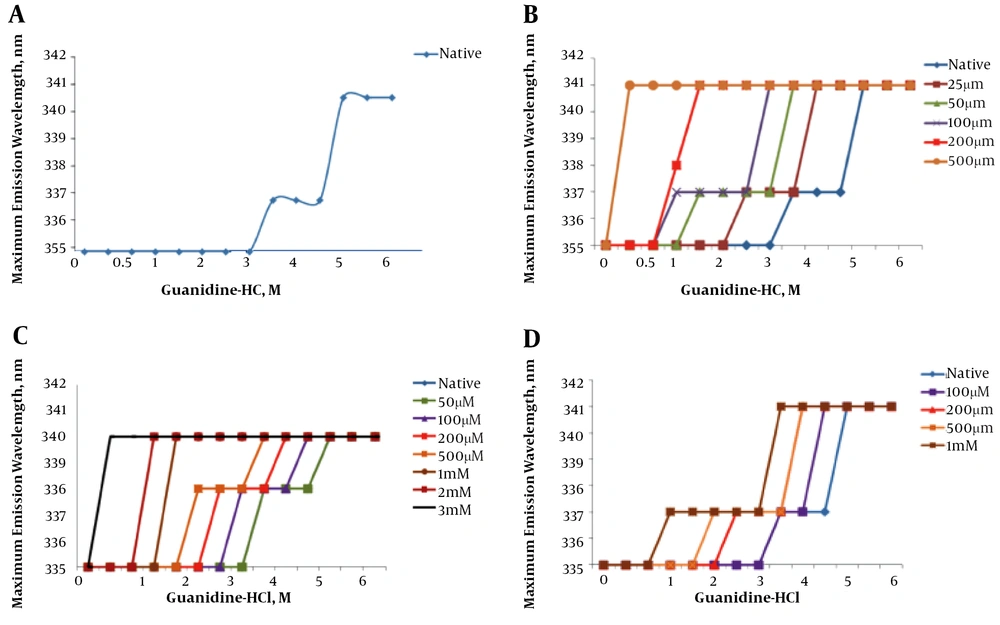
Figure 4A demonstrates the effect of increasing concentrations (0 to 6 M) of guanidine hydrochloride on the wavelength of maximum intrinsic fluorescence observed for the hFGFR2b protein. The graph shows the fragment unfolds as a sigmoidal curve with two transitions, suggesting that each domain was unfolded independently.
Chemical induced denaturation of FGFR2b kinase domain. A, Represents native spectra and B, C, D, and E represents unfolding in presence gallic acid (from 25 µM to 500 µM), naringenin (from 50 µM to 3 M), quercetin (from 100 µM to 1 M) and catechin (from 100 µM to 500 µM), respectively. The graphs represent the unfolding of the kinase domain structure in the presence of flavonoids and increasing concentrations of guanidine hydrochloride at room temperature.
4.4. Chemical Induced Denaturation of FGFR2b Kinase Domain in the Presence of Increasing Concentration of Gallic Acid
Figure 4B shows a step-wise chemical denaturation of FGFR2b in the absence and the presence of increasing concentration of gallic acid (from 25 µM to 500 µM). The graph clearly demonstrates that an increase in concentration of gallic acid leads to a significant change in the unfolding curve of the FGFR2b kinase domain, significantly shifting the midpoints of the transition states of FGFR2b kinase domain to the left. This finding suggests that presence of the flavonoid destabilizes the tertiary structure of the kinase protein.
4.5. Chemical Induced Denaturation of FGFR2b Kinase Domain in the Presence of Increasing Concentration of Naringenin
Figure 4C shows the step-wise chemical denaturation of FGFR2b in the absence and the presence of increasing concentration of naringenin (50 µM to 3 mM). The graph clearly demonstrates that an increase in concentration of naringenin results in a considerable change in the unfolding curve of the FGFR2b kinase domain, remarkably shifting the midpoints of the transition states FGFR2b kinase domain to the left. This finding suggests the presence of naringenin impinging on the tertiary structure of the kinase protein.
4.6. Chemical Induced Denaturation of FGFR2b Kinase Domain in the Presence of Increasing Concentration of Quercetin
Figure 4D shows a step-wise chemical denaturation of FGFR2b in the absence and the presence of increasing concentration of quercetin (100 µM to 1 mM). The graph clearly demonstrates that an increase in concentration of quercetin causes a remarkable change in the unfolding curve of the FGFR2b kinase domain, considerably shifting the midpoints of the transition states FGFR2b kinase domain to the left. This finding suggests that the presence of quercetin destabilizes the tertiary structure of the kinase protein.
4.7. Chemical Induced Denaturation of the FGFR2b Kinase Domain in the Presence of Increasing Concentration of Catechin
Figure 4E shows a step-wise chemical denaturation of FGFR2b in the absence and presence of increasing concentration of catechin (100 µM to 500 µM). The graph clearly demonstrates that an increase in concentration of catechin causes a significant change in the unfolding curve of the FGFR2b kinase domain, significantly shifting the midpoints of the transition states FGFR2b kinase domain to the left. This finding suggests that presence of catechin destabilizes the tertiary structure of the kinase protein.
5. Discussion
In this study, to express an active hFGFR2b kinase domain, an expression vector containing an engineered (mutated) coding region of hFGFR2b kinase domain was used (23, 24). The vector had two gain-of-function mutations created in (E565A) and (K660E) residues. These mutations were selected based on the associated pathologic disorders, namely Pfeiffer syndrome (25) and endometrial cancer (26, 27), respectively. The recombinant kinase domain was over-expressed in E. coli expression system and purified using a Ni-NTA affinity chromatography (28, 29). The PAGE analyses were then performed to evaluate whether or not the kinase domain was in active conformational state. To this end, interaction of the kinase domain with both the wild type nSH2-cSH2 (as a positive control) and the mutated nSH2-cSH2 (as a negative control; the specific mutations were included in nSH2-cSH2 so that its interaction site with the kinase domain was adversely affected) of PLC-γ, were assessed. From the PAGE data, it appeared that the purified kinase domain interacted with the wild type SH2 domains yet not with the mutant SH2 domains of PLC-γ,. This is evident by the fact that the wild type nSH2-cSH2 and the kinase domain formed a complex, appearing as a single band on the PAGE gel, whereas the mutant nSH2-cSH2 domains and the kinase domain did not form a complex, thus did not appear as a single band on the PAGE gel. This strongly demonstrated that the kinase domain was in active conformational state. The effect of four flavonoids, gallic acid, naringenin, quercetin and catechin on structural stability of the recombinant kinase domain was then studied (30-33).
The fluorescence analysis revealed that the overall structure of hFGFR2b kinase domain was destabilized upon interaction with gallic acid, naringenin, quercetin and catechin. This is evidenced by the fact that increasing concentrations of gallic acid, naringenin, quercetin, and catechin induced a significant red-shift in the wavelength of maximum emission. As previously explained, the kinase domain of FGFR2b consists of two lobes, the N-terminal and the C-terminal lobes (34, 35). In line with this, the chemical-induced unfolding analysis clearly showed that the kinase domain was unfolded as a sigmoidal curve with two transitions, thus, suggesting the two lobes of the domain was unfolded independently. Importantly, comparisons of unfolding curves of the kinase domain in the presence and absence of gallic acid, naringenin, quercetin, and catechin revealed that the flavonoids destabilized both lobes of the kinase domains, impinging on overall structure of the FGFR2b kinase domain. This is evident by the chemical-induced unfolding data, showing that an increasing concentration of either of the flavonoids considerably shifted the midpoint transitions to the left (from 3.25 M and 4.75 M to lower concentration of guanidine hydrochloride). These data suggested that flavonoids reduced the activity of the FGFR2b kinase domain and thereby down-regulated FGF signaling. Consistent with these findings, several in vitro and cell-based studies have suggested that some flavonoids and unsaturated fatty acid modulate cellular signaling, acting as cancer chemo-preventative agents (36-38). As an example, Hou et al. (3) suggested that some flavonoids directly interact with signaling protein kinases, such as Akt/protein kinase B (Akt/PKB), Fyn, Janus kinase 1 (JAK1), mitogen-activated protein kinase 1 (MEK1), phosphoinositide 3-kinase (PI3K), mitogen-activated protein (MAP) kinase kinase 4 (MKK4), Raf1, and zeta chain-associated 70-kDa protein (ZAP-70) kinase, acting as anti-proliferative agent (20). Additionally, several cell-based analyses have also suggested that flavonoids act as antioxidants, controlling multiple cellular signaling pathways (3, 39). For instance, a comprehensive cell-based study suggested that flavonoids bind to signaling protein kinases, regulating expression of some antioxidant enzymes, such as HO-1, NQO-1 and GSH (12). Taken together, the current findings together with previous in vitro and cellular studies suggest that interaction of flavonoids with signaling protein kinases, such as FGFR2b, imping on active conformations of signaling molecules, thus down-regulating their kinase activities. These results therefore provide a possible molecular mechanism, by which flavonoids exert their anti-proliferative functions, thus, suggesting that they may have a place as adjuvant therapy in combination with tyrosine kinase inhibitors (TKIs).