1. Background
Acute lymphoblastic leukemia (ALL) is a hematological malignancy affecting B or T lymphoid cell precursors. The current standard treatment for ALL is based on chemotherapy, bone marrow transplantation, and radiation therapy for central nervous system (CNS) protection (1). However, there is a need for new therapeutic approaches as a significant number of patients have poor prognosis and do not respond appropriately to existing therapies (2).
Autophagy and apoptosis are crucial in normal functioning and developing diseases like cancer. Apoptosis plays a critical role in regulating cancer cell death. At the same time, autophagy acts as a protective mechanism by safeguarding healthy cells against genetic damage, oxidative stress, and excessive stem cell proliferation (3, 4). Both autophagy and apoptosis are involved in ALL recurrence (5). Extensive research on the cross-talk between autophagy and apoptosis has shown that autophagy functions in parallel with apoptosis, either promoting cell survival by repressing apoptosis or acting as a prerequisite for apoptosis (6). Several conserved genes, including MAP1LC3, P62/SQSTM1, Atg2B, and DRAM1, have been identified as important regulators of the molecular mechanism underlying autophagy (7). Various transcription factors, such as FOXO, STAT1, STAT3, NF-κB, and TP53, modulate the expression of these genes. In addition, post-transcriptional effectors, including microRNAs, siRNAs, long non-coding RNAs (lncRNAs), and modifications such as phosphorylation, ubiquitination, acetylation/deacetylation, and histones acetylation are critical in this regulation (8, 9).
Extensive investigations have shown the effectiveness of natural compounds in cancer therapy, particularly through the induction of apoptosis and autophagy (10). One such compound is epigallocatechin gallate (EGCG), a polyphenol found in green tea. It is the most important catechin leading to apoptosis, proliferation arrest, and autophagy induction or inhibition in cancer cells (11). Numerous in vitro and in vivo studies have demonstrated that EGCG does not harm normal cells (12, 13). Epigallocatechin gallate exhibits a multifaceted mechanism of action in leukemic cells. It inhibits key signaling pathways like PI3K/Akt and MAPK, modulates oxidative stress and inflammation, regulates transcription factors and miRNAs, and influences cell cycle proteins such as CDKs. Furthermore, EGCG can modulate epigenetic processes (14), disrupt leukemic cell-microenvironment interactions (15), and enhance the sensitivity of leukemic cells to chemotherapy (16). Several clinical trials have investigated the mechanism, efficacy, safety, and tolerability of EGCG alone or in combination with chemotherapeutic drugs for the various types of leukemia treatment (17).
There are conflicting findings regarding the relationship between autophagy and apoptosis in NALM-6 cell line. Some studies have suggested that blocking autophagy causes cell death (18, 19), while others have reported a decrease in cell death when autophagy is inhibited (20, 21). Additionally, evidence shows that inhibiting autophagy does not affect cell viability significantly in this specific cell line (22).
2. Objectives
The ability of EGCG in apoptosis induction and autophagy induction or inhibition without harming healthy cells has made it an attractive candidate for new investigations in the development of potential cancer treatments. Given the conflicting data and the importance of autophagy and apoptosis cross-talk in treating ALL, this study aims to explore the effect of EGCG on autophagy, apoptosis, and their interaction in NALM-6 cell line. It provides original insights into the potential use of EGCG as a novel treatment for ALL. The research will contribute to the existing knowledge and shed light on the complex mechanisms underlying the effectiveness of EGCG in ALL treatments, paving the way for further advancements.
3. Methods
3.1. Cell Culture
NALM-6, a pre-B-ALL cell line, was purchased from the National Cell Bank of Iran (Pasteur Institute, Iran). They were maintained in RPMI1640 media, supplemented with 10% fetal bovine serum, 100 IU/mL penicillin, 0.1 mg/mL streptomycin (Life Technologies, USA), and 2 mM L-glutamine (Shell Max, China) in an environment of 5% CO2 and humidified atmosphere of 95% at 37°C.
3.2. 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl Tetrazolium Bromide Assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay was employed to measure the impact of EGCG on the cell viability of NALM-6. The cells were seeded at a density of 1 × 104 cells per well in 96-well cell culture plates in 100 µL of growth medium and incubated for at least 2 hours. EGCG (Abcam, UK) was dissolved in dimethyl sulfoxide (DMSO) (Shellmax), and different concentrations of EGCG (ranging from 2 to 110 µM) were prepared by diluting the stock solution in the culture medium. The cells were exposed to the EGCG solutions and incubated for 24 and 48 hours. To assess cell viability, 10 µL of a sterile 5 mg/mL solution of 3-4, 5-dimethylthiazol-2-yl-2, 5-diphenyl-tetrazolium bromide (Melford, UK) was added to each well. After 4 hours of incubation, the formazan product was solubilized using DMSO (Merck KGaA, Germany), and spectrometric absorbance was measured at 545 nm using a microplate photometer (Stat Fax 2100, USA). Three independent experimentations were conducted in quadruplicate to ensure the reliability of the results.
3.3. Trypan Blue Exclusion Assay
During our experiments, the auto-oxidation of EGCG in culture conditions caused a brown color, interfering with the MTT assay as a colorimetric method. This problem was resolved using no-cell control wells to account for any background color; still a trypan blue exclusion assay was performed to confirm the outcomes of the MTT assay. Briefly, the cells were seeded at a density of 1 × 105 cells per well in 24-well plates in 700 µL of growth medium and left to incubate for a minimum of 2 hours. The cells were exposed to various doses of EGCG (2 - 110 μM) for 24 and 48 hours. The cells were then washed and resuspended in 1X phosphate-buffered saline (PBS) containing 0.4% trypan blue (Sigma-Aldrich, USA) and counted using a hemocytometer following the standard protocol. Two separate experiments were conducted in duplicate.
3.4. Flow Cytometry for Apoptosis
To investigate the effect of EGCG on apoptosis, the cells were seeded at a density of 1 × 105 cells per well in 24-well plates in 700 µL growth medium and incubated for at least 2 hours. They were then treated with different concentrations of EGCG (2 - 110 µM) for 24 and 48 hours. In addition, to indicate the cross-talk between autophagy and apoptosis, some cells were treated with EGCG 45 µM alone and some in combination with NH4Cl 10 mM during the last 12 hours of 36-hour incubation. Apoptotic cells were assessed using PE-Annexin V/7-AAD Apoptosis Detection Kit I (BD Life Sciences, USA). After washing with 1X PBS, the cells were suspended in 100 µL of 1X Annexin V binding buffer. They were stained with PE-Annexin V and 7-AAD for 15 minutes in a dark room. Subsequently, 400 µL of Annexin V binding buffer was added, and the cells were evaluated using a FACSCalibur flow cytometer (BD Life Sciences) within 1 hour. Two independent experiments were performed, and data were analyzed by FlowJo™ v10.5.4 software (BD Life Sciences).
3.5. Western Blotting
Five million NALM-6 cells were cultured until they reached 70 - 80% confluency. Subsequently, they were treated with EGCG at concentrations of 20 µM and 45 μM for 36 hours. During the last 12 hours of incubation, NH4Cl 10 mM (Merck KGaA), dissolved in sterile deionized water, was added to inhibit autophagy. This compound impedes autophagic flux by raising the luminal PH of intracellular vesicles and preventing degradative enzyme activation inside lysosomes. After the treatment, the cells were rinsed once with PBS and subsequently lysed using ice-cold lysis buffer with a 1X formulation. The lysis buffer contained Tris-HCl (0.06 M, pH 6.8), sodium dodecyl sulfate (SDS) (2%), bromophenol blue (0.2%), glycerol (20%), and was supplemented with a protease phosphatase inhibitor cocktail from Sigma-Aldrich. The resulting protein collection was obtained by centrifugation at 4°C for 30 minutes. Equal aliquots of 20 μg protein, assessed using the Bradford method, were boiled in 5X loading buffer containing Tris-HCl (0.06 M, PH 6.8), SDS (2%), bromophenol blue (0.2%), and glycerol (20%) at 90°C for 5 minutes. The samples were then dissolved using SDS-polyacrylamide gel electrophoresis (PAGE) on 12% gels. Next, they were transferred to polyvinylidene fluoride (PVDF) from Roche for 1 hour and blocked for 1 hour with 5% skim milk in tris-buffered saline with 0.1% tween 20 detergent (TBS-T). The blots were probed with a rabbit anti-LC3 A/B antibody (1:1000) from cell signaling technology and a mouse anti-β-actin antibody (1:1000) from Santa Cruz biotechnology in 5% non-fat skim milk in TBS-T at 4°C overnight. After washing with TBS-T, the blots were treated with horseradish peroxidase-conjugated anti-rabbit IgG from Sigma-Aldrich and anti-mouse IgG from Bio-Rad for 1 hour. They were then washed with TBS-T three times for 5 minutes. Finally, the proteins were detected using an enhanced chemiluminescence system (ChemiDoc™ MP System, Bio-Rad, USA) and analyzed using Image J version 1.52 software (National Institutes of Health, USA). Three experiments were performed in triplicate to ensure the reproducibility of the results.
3.6. Real-time PCR
NALM-6 cells were seeded at a density of 1.2 × 106 in 3.3 mL of the complete medium in 6-well plates and treated with EGCG 45 µM for 36 hours to examine its effect on the expression of autophagy-related genes. Total RNA was extracted from the cells using TRI reagent (Sigma-Aldrich) according to the manufacturer′s instructions, and the RNA concentration was measured using a NanoDrop instrument (Hellma NY, China). Reverse transcription was performed using 500 ng of total RNA and the Prime Script TM RT reagent Kit (Yektatajhiz Azma, Iran) to synthesize complementary DNA (cDNA). Quantitative real-time PCR (qRT-PCR) was carried out on a Rotor gene system (Qiagen, Germany) using an SYBR Premix Ex Taq (Yektatajhiz Azma). The reaction mixture with a total volume of 20 µL consisted of 8 µL DNase free water, 0.5 µL of 10 pmol/µL forward and reverse primers, 10 µL of SYBR Premix Ex Taq, and 1 µL of cDNA template. The experimental conditions were based on the PCR program presented in Table 1. The β-2 microglobulin gene was used as an internal control for the relative quantification of gene expression. As part of the qRT-PCR, a negative control was incorporated using a no-template sample. Besides, a positive control RNA extracted from starved cells was included. The comparative relative quantification of gene expression was done using the Pfaffl method, with data from three independent experiments conducted in triplicate. The primers for P62/SQSTM1, Atg2B, LC3B, and DRAM1 genes (Table 2) were designed using the Allele ID v7.70 software (PREMIER Biosoft, USA).
| Step | Temperature (°C) | Time (sec) | Cycling |
|---|---|---|---|
| Holding | 95 | 90 | 1 |
| Denaturing | 95 | 5 | 40 |
| Annealing | 57 | 30 | |
| Extension | 72 | 30 |
| Gene and Primer Name | Sequence (5′-3′) |
|---|---|
| P62/SQSTM1 | |
| Forward | 5′-CTTTGTAGCCATCCTGTTA-3′ |
| Reverse | 5′-CGACTATGTGACCTCTTTAT-3′ |
| LC3B | |
| Forward | 5′-GTGATAATAGAACGATACAAGG-3′ |
| Reverse | 5′-CACTCTCATACACCTCTG-3′ |
| DRAM1 | |
| Forward | 5′-AAGGAATGACTAAGGATGTT-3′ |
| Reverse | 5′-CAGGCGATACAGACTATT-3′ |
| Atg2B | |
| Forward | 5′- CCAGAAACTAACAGATGAACAAG-3′ |
| Reverse | 5′-TCAGCAGTTTCATCACAGTACAC-3′ |
| β2m | |
| Forward | 5′-TATGCCTGCCGTGTGAAC-3′ |
| Reverse | 5′-CTACCTGTGGAGCAACCTG-3′ |
3.7. Statistical Analysis
To ensure accurate comparisons, control groups in all experiments were not exposed to any chemicals. These control groups served as a baseline to compare with the experimental groups treated with chemicals. Each examination was repeated appropriately to provide an adequate sample size. Statistical analyses were performed using GraphPad Prism v 8.4.3 software (Insightful Science, USA). The data were represented as mean ± SD, two-way ANOVA, and unpaired t-tests were employed to analyze the data. In the case of two-way ANOVA, post hoc tests such as Tukey and Fisher′s LSD were used to determine significant differences between groups. The significance of the data was determined by a P-value below 0.05.
4. Results
4.1. EGCG Decreases Cell Viability of NALM-6 Cell Line
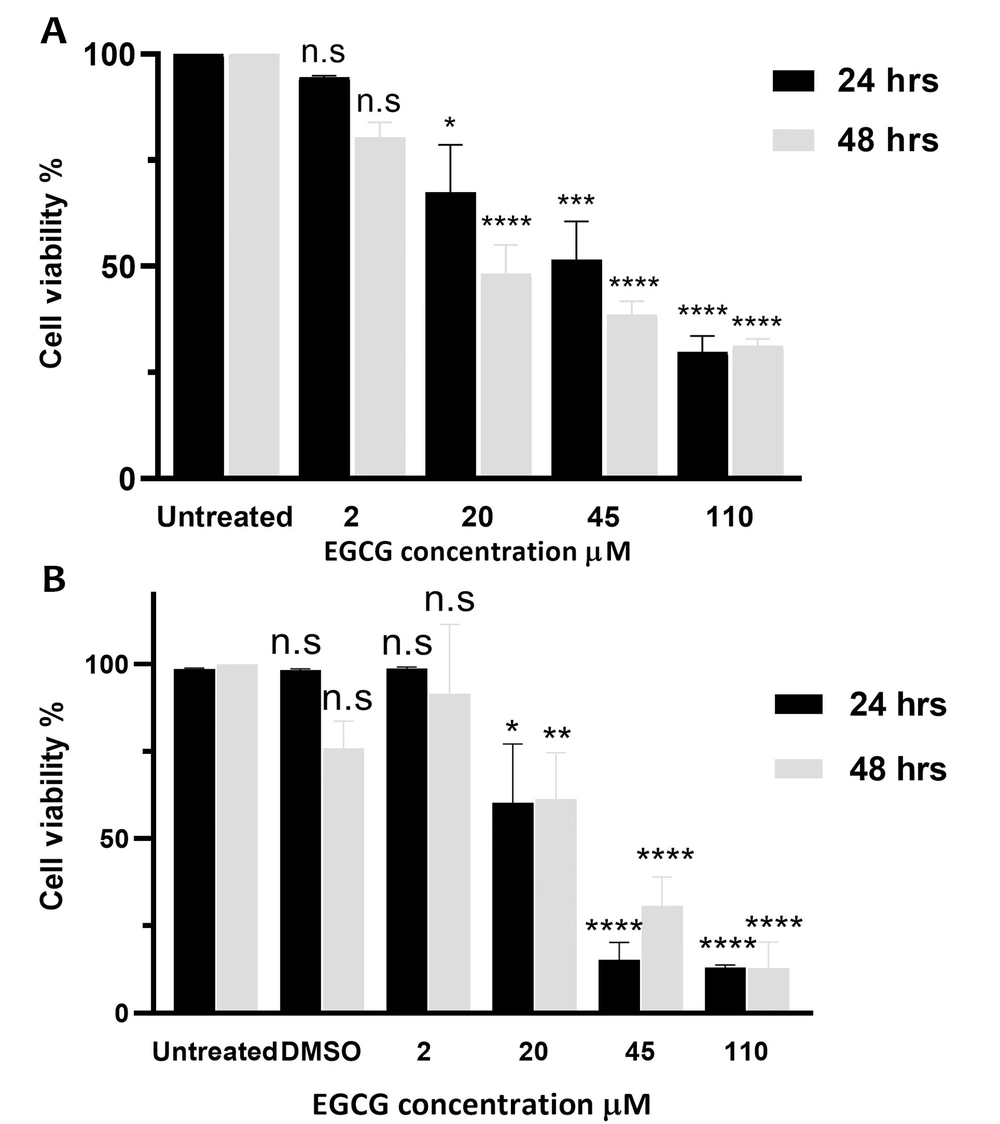
The MTT assay assessed the impact of varying doses of EGCG (2 - 110 μM) on proliferation and cell viability. The findings showed that cell viability was significantly decreased by 5.4 ± 0.5%, 32.6 ± 19.4%, 48.5 ± 15.6%, and 70.2 ± 6.6% after 24 hours of exposure to EGCG 2, 20, 45, and 110 µM, respectively. Furthermore, following 48 hours of treatment with EGCG at concentrations of 2, 20, 45, and 110 µM, the cell viability reduced by 19.6 ± 6%, 51.8 ± 11.8%, 61.4 ± 5.4%, and 68.7 ± 2.7%, respectively (Figure 1A) (P < 0.0001). However, no significant difference was observed between the outcomes obtained after 24 and 48 hours of incubation (P > 0.05).
Epigallocatechin gallate (EGCG) decreases proliferation and cell viability in NALM-6 cell line. EGCG at concentrations ranging from 2 to 110 µM was administered to NALM-6 cells for 24 and 48 hours. Graph A, displays the mean ± SD of three independent 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays conducted in quadruplicate; and graph B, shows the mean ± SD of two independent trypan blue exclusion assays performed in duplicate. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. n.s, non-significant.
4.2. EGCG Induces Cell Death in NALM-6 Cell Line
The results of the trypan blue exclusion assay indicated that after 24 hours, EGCG 2 µM, 20 µM, 45 µM, and 110 µM led to 1.2 ± 0.4%, 39.8 ± 16.9%, 84.7 ± 5.0%, and 87.0 ± 0.3% cell death, respectively (P < 0.0001). Similarly, treatment with these concentrations of EGCG for 48 hours caused 8.5 ± 1.7%, 38.6 ± 13.2%, 69.3 ± 8.2%, and 87.2 ± 7.6% cell death, respectively (P < 0.0001). Overall, EGCG significantly induced cell death in a dose-dependent but not time-dependent manner (P > 0.05) (Figure 1B).
4.3. EGCG Induces Apoptosis in NALM-6 Cell Line
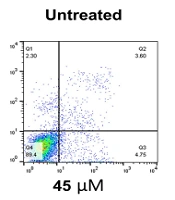
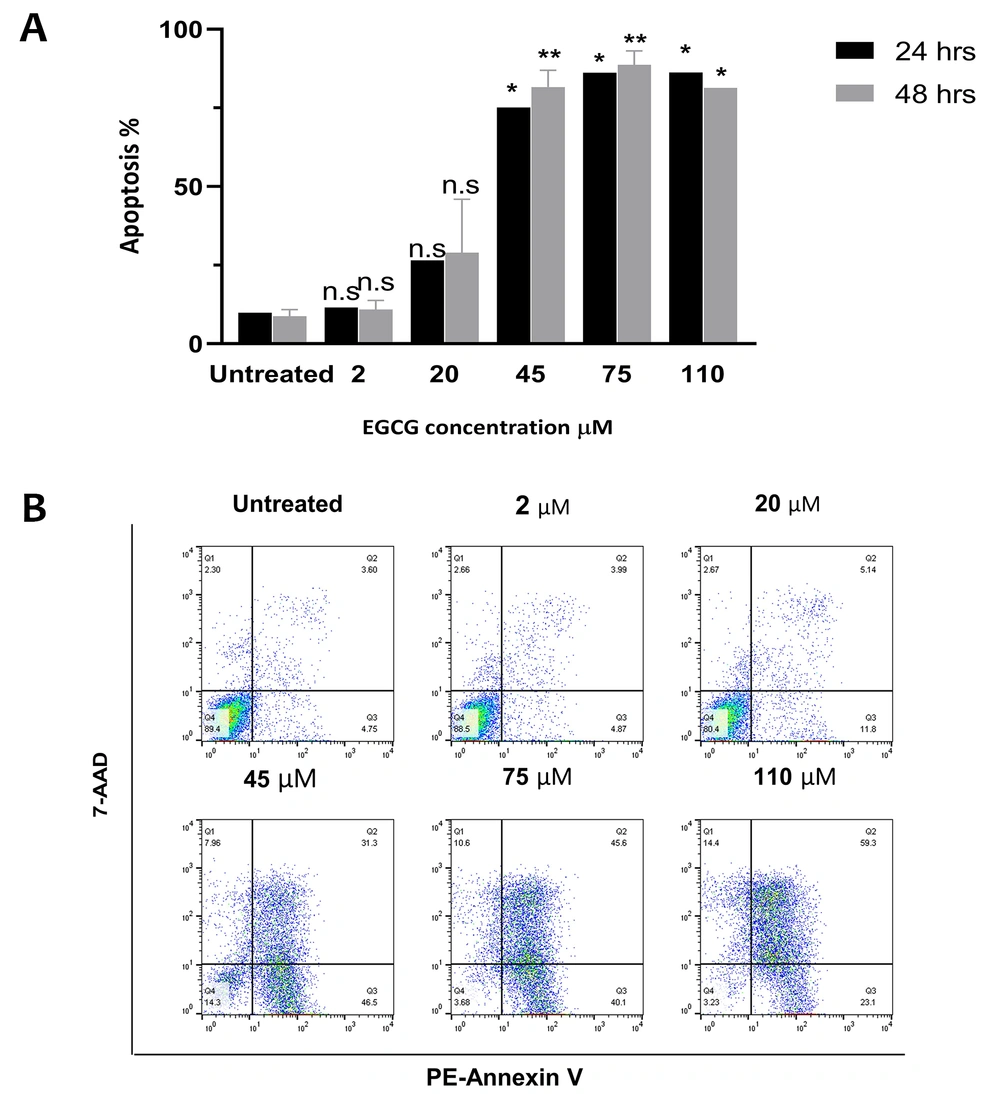
Flow cytometry analysis was carried out to assess the EGCG effect on apoptosis in NALM-6 cell line. Our findings showed that treatment with 20 µM EGCG resulted in 16.7% apoptosis after 24 hours, which increased to 20.2% after 48 hours. It was also observed that higher concentrations of EGCG prompted a greater percentage of apoptosis. Specifically, the data revealed that after 24 hours of treatment, EGCG 45 µM led to apoptosis rates of 65.2% and 72.8%, whereas at 75 µM the apoptosis rates were 76.3% and 79.9% after 48 hours. Additionally, EGCG caused over 80% apoptosis at both time points (P = 0.0003), and there was no significant difference between the data obtained after 24 and 48 hours of incubation (P > 0.05) (Figure 2).
Epigallocatechin gallate (EGCG) induces apoptosis in NALM-6 cell line. EGCG at concentrations 2 - 110 µM was administered to the cells for 24 and 48 hours. Graph A, represents the data as the mean ± SD of two independent flow cytometric-based experiments; graph B, depicts annexin V-PEneg7-AADneg cells in the lower left quadrant, considered live cells, and annexin V-PE+-7-AADneg cells in the lower right quadrants, indicating apoptotic cells. * P < 0.05, ** P < 0.01. n.s, non-significant.
4.4. EGCG Induces Autophagy in NALM-6 Cell Line
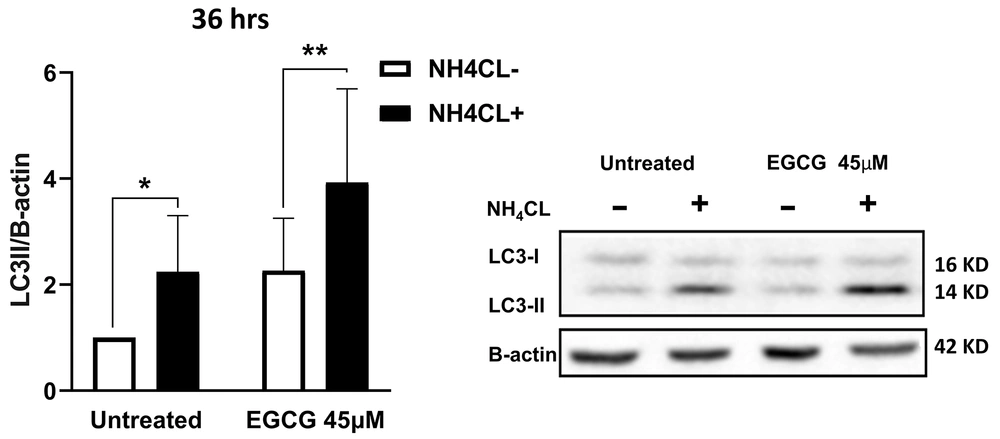
To investigate the effect of EGCG on autophagy, NALM-6 cells were treated with two different concentrations of EGCG (20 µM and 45 μM) for 36 hours. Western blot analysis was used to monitor the LC3 protein level, a commonly used marker for autophagy. Our findings showed that treatment with EGCG 45 µM caused a higher conversion of LC3-I to LC3-II than 20 µM within 36 hours (presented in Supplementary File, Appendix 1). These findings suggest that higher concentrations of EGCG trigger a more pronounced activation of autophagy. The cells were subsequently treated with NH4Cl 10 mM to investigate whether the increase in LC3 level was due to an impairment of autophagic flux or activation of autophagy. The findings revealed a significant accumulation of LC3-II after 12 hours of treatment with NH4Cl. Notably, LC3-II expression in the treated cells with EGCG 45 µM was twice as much as in the untreated cells (P = 0.001), while added NH4Cl led to more than a four-fold increase, suggesting that activation of autophagy, rather than flux impairment, is responsible for the increased levels of LC3-II upon EGCG treatment (P = 0.001) (Figure 3). Based on these findings, EGCG at a concentration of 45 µM was selected as the optimal concentration for subsequent experiments, as it induced significant autophagy activation without causing complete flux impairment.
Epigallocatechin gallate (EGCG) induces autophagy in NALM-6 cells. EGCG 45 µM was administered to the cells for 36 hours, with the presence or absence of NH4Cl at 10 mM during the last 12 hours. The data represented the mean ± SD of three independent western blotting experiments in triplicate. * P < 0.05, ** P < 0.01.
4.5. EGCG Affects the Autophagy Genes Expression
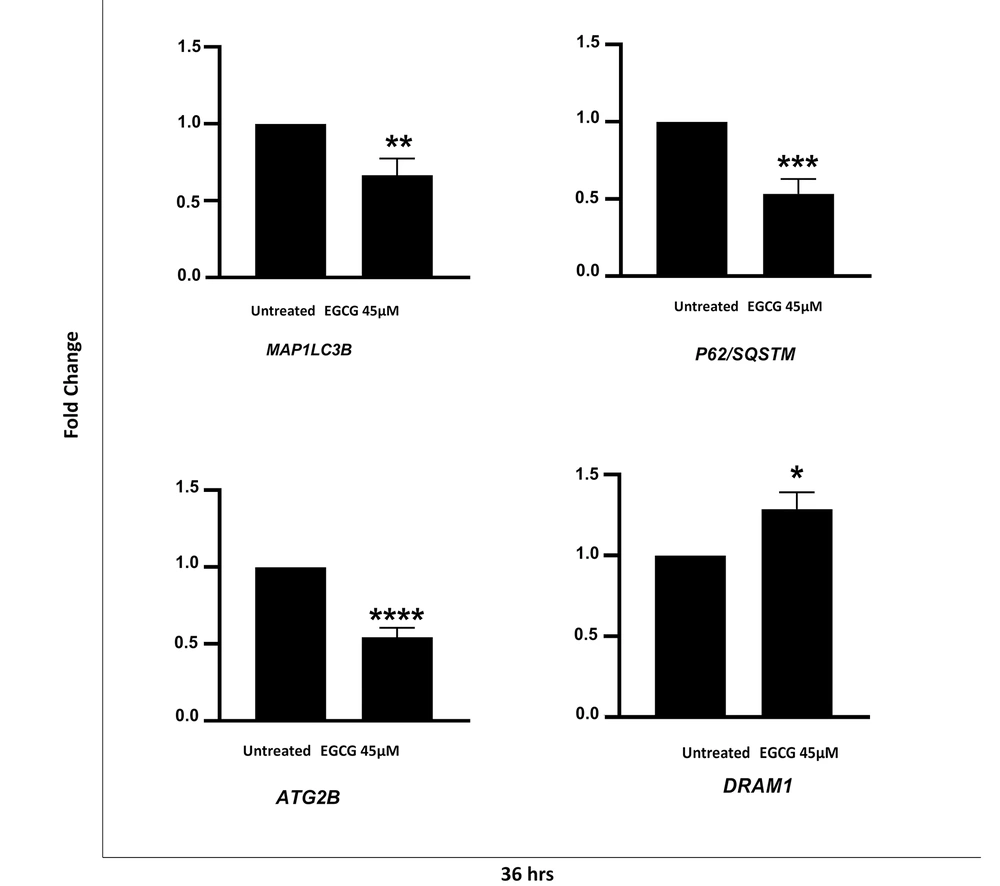
To further investigate whether the EGCG treatment affected the autophagy at a transcriptional level, qRT-PCR was performed to evaluate the level of LC3B, P62/SQSTM1, Atg2B, and DRAM1 genes following EGCG 45µM treatment. Our data showed that EGCG led to a significant reduction in the expression of LC3B, P62, and Atg2B genes by 33.3% (P = 0.008), 46.5% (P = 0.0001), and 45.5% (P ≤ 0.0001), respectively. We also observed a 1.3-fold increase in DRAM1 expression following the EGCG 45 µM treatment (P = 0.013) (Figure 4).
Epigallocatechin gallate (EGCG) alters autophagy-related gene expression in NALM-6 cell line. EGCG 45 µM was administered to the cells for 36 hours. Data represented as mean ± SD of three independent real-time PCR experiments in triplicate. * P < 0.05, ** P < 0.01, *** P < 0.001 **** P < 0.0001.
4.6. Inhibition of Autophagy Does Not Interfere with Apoptosis in NALM-6 Cell Line
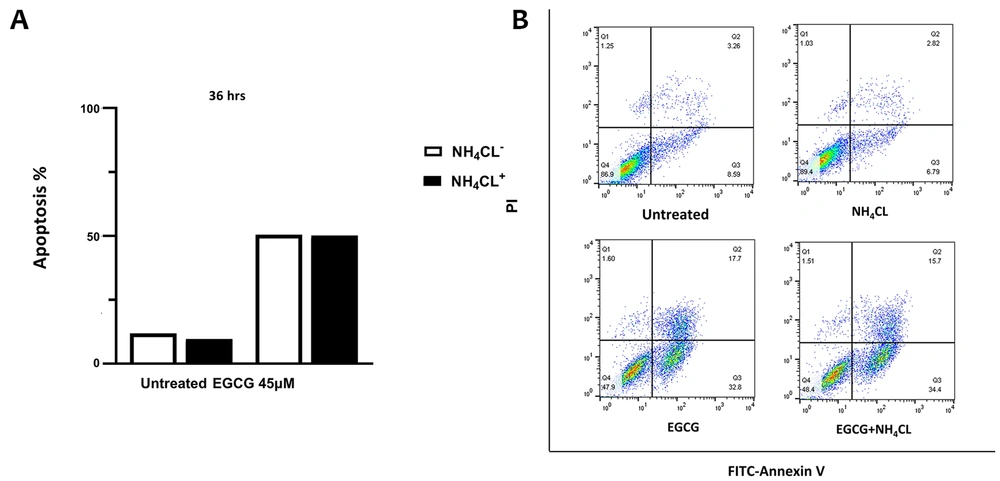
Flow cytometry analysis was employed to investigate the relationship between autophagy and apoptosis in NALM-6 cells. EGCG 45 μM was administered to the cells for 36 hours, with the addition or omission of NH4Cl 10 mM during the last 12 hours of incubation. It can be inferred from Figure 5 that the blockage of autophagy with NH4Cl did not substantially affect the apoptosis rate in either the untreated or EGCG-treated cells.
NH4Cl does not affect apoptosis in epigallocatechin gallate (EGCG)-treated NALM-6 cells. The cells were treated with EGCG 45 µM for 36 hours and with or without NH4Cl 10 mM during the final 12 hours. Graph A, displays the rate of apoptotic cell death when NH4Cl is present or absent; graph B, the lower left quadrant represents live cells, characterized by annexin V-FITCneg PIneg, and the lower right quadrants show annexin V-FITC+-PIneg cells, including apoptotic cells.
5. Discussion
Despite significant improvements in chemotherapy outcomes for pediatric ALL patients, the prognosis for adults and infants remains poor (23). Identifying the potential of natural compounds with the slightest toxicity and strong efficacy is crucial for developing innovative therapies that could enhance the effectiveness of currently available treatments. Although the impact of EGCG on cell viability or apoptosis in leukemic cell lines has been extensively examined previously, little evidence exists regarding its effect on autophagy or the cross-talk between autophagy and apoptosis in leukemia. In this study, we focused on B lymphoblastic leukemia, aiming to explore how EGCG affects autophagy, apoptosis, and their intricate relationship in NALM-6 cell line.
Our findings indicated a significant decrease in cell viability following treatment with varying concentrations of EGCG at both 24 and 48 hours. There was no significant difference in the results obtained after these time points. These findings align with previous studies that have demonstrated the inhibitory effects of EGCG on proliferation and its ability to DNA fragmentation in various human leukemic cell lines in a dose-dependent manner (24). In addition, we revealed that EGCG induced apoptosis in NALM-6 cells in a dose-dependent but not time-dependent manner. Treatment with 20 µM EGCG for 24 hours resulted in 16.7% apoptosis, which increased to 20.2% after 48 hours. Interestingly, higher concentrations of EGCG led to a greater rate of apoptosis. Our findings are consistent with a study by Yin et al., which has reported that EGCG treatment led to apoptosis in bladder cancer cell lines in a dose-dependent manner (25). Furthermore, Cornwall et al. have demonstrated that EGCG-induced apoptotic cell death in B-CLL and T-CLL cells is dose-dependent while leaving normal B and T cells unaffected (26). Della Via et al. have reported a significant increase of apoptotic cells in the spleen of PML/RARα mice (27). Given the intricate relationship between autophagy and apoptosis pathways (28) and the dual role of autophagy in cancer pathophysiology (4), several studies have investigated the influence of EGCG on either apoptosis or autophagy pathways, as well as their cross-talk. Satoh et al., in an in vitro study on five different mesothelium cell lines, found that EGCG prompted apoptosis and autophagy, while inhibition of autophagy using chloroquine led to cell death in EGCG-treated cells (29). It has also been indicated that EGCG mitigated the toxicity of bortezomib by inducing autophagy and safeguarding PC3, a prostate cancer cell line, from death (30). In our study, Western Blot analysis for LC3-II levels showed that treatment with EGCG 45 µM effectively stimulated autophagy in NALM-6 cell line, and the increase in LC3-II levels was due to the activation of autophagy rather than an impairment of autophagic flux. However, autophagy inhibition by NH4Cl did not affect apoptosis in the presence or absence of EGCG. Our data are consistent with Wong et al. study, which has demonstrated that autophagy suppression had no effect on apoptosis in NALM-6 cell line (22), contrary to others who either revealed that inhibition of autophagy increased cell death (18, 19) or could protect this particular cell line (21). These results suggest that the relationship between autophagy and apoptosis is complex and may vary depending on a specific context and cell type.
Furthermore, we observed a significant reduction in the mRNA level of LC3 in NALM-6 cells treated with 45 µM of EGCG for 36 hours, compared with the untreated cells. These findings align with Li et al.′s study, which reported that EGCG 20 µM triggered a significant decrease in LC3 mRNA levels in the liver tissue of animal models pretreated with concanavalin A, an acute hepatitis inducer (31). It can be suggested that decreasing LC3 mRNA level may be related to the suppressing of the FOXO1 transcription factor by EGCG (32). In contrast to the findings of Zhong et al. in HepG2 cells (a hepatocarcinoma cell line), our study revealed a significant reduction in P62 mRNA expression in NALM-6 cells following the treatment with EGCG 45 µM (33). Nonetheless, in another study, DIV8 primary rat cortical neurons treated with EGCG at a concentration of 50 μM for 24 hours exhibited a notable rise in P62 mRNA levels compared to DMSO control (34). The susceptibility of P62, as one of the Nf-ƘB target genes, to inhibition by EGCG suggests the capacity of this compound to reduce P62 expression (35).
In contrast to LC3B and P62 genes, no evidence exists regarding the influence of EGCG on Atg2B and DRAM1 genes. We found a significant reduction in ATG2B mRNA expression in NALM-6 cells in response to the treatment with EGCG 45µM after 36 hours. However, Atg2B gene expression remained constant in MEFs (mouse embryonic fibroblast cells) after 8 hours of starvation (36). Additionally, it has been noted that hepatic cell lines such as 7702, HepG2, Hep3B, and Huh7 cells underwent DRAM-1 mediated autophagy after 48 hours of starvation (37). Our findings also demonstrated that treatment of NALM-6 cells with EGCG at a concentration of 45 µM led to a modest yet noteworthy enhancement in DRAM1 gene expression.
Our results suggest that the decrease in mRNA levels of autophagy genes and the increase in DRAM-1 gene expression may be related to apoptosis induction by EGCG 45 µM. On the other hand, the observed downregulation of autophagy genes and induction of autophagy following EGCG treatment in NALM-6 cells may be ascribed to post-transcriptional or post-translational modifications. Therefore, further inquiries are warranted to delineate the precise underlying mechanism through which EGCG modulates the expression of these genes.
5.1. Conclusions
In conclusion, our findings revealed that EGCG reduces cell viability and results in apoptosis in NALM-6 cell line in a dose- but not time-dependent manner. In addition, EGCG induces autophagy dose-dependently and an alteration in LC3B, P62/SQSTM1, Atg2B, and DRAM1 gene expression. Nevertheless, the inhibition of autophagy does not interfere with the apoptotic cell death triggered by EGCG. Considering the significance of autophagy as a secondary cell death mechanism, it would be of great importance to further investigate the possible involvement of EGCG in inducing autophagic cell death in cells in which the apoptotic pathway is impaired. Recently, several clinical trials in phase I or II studying the effect of EGCG on leukemic patients have highlighted the potential clinical utility of EGCG as a targeted therapy for ALL patients, particularly in cases where current treatments have limited efficacy (38, 39). Therefore, new clinical studies are required to validate our findings and determine the optimal dosage and treatment regimen for EGCG in ALL patients, especially in combination with chemotherapeutic drugs. These studies will provide valuable insights into the feasibility of incorporating EGCG into standard treatment protocols, potentially improving the response to currently available therapies and ultimately enhancing treatment outcomes in ALL.