1. Context
1. Type 2 Diabetes Mellitus and Its Relation with α-glucosidase
Type 2 diabetes mellitus (T2DM) is a chronic endocrine metabolic disorder characterized by insulin resistance, impaired glucose regulation, and a wide array of metabolic dysfunctions. Due to changes in social and cultural lifestyle, as well as economic development, the global prevalence of diabetes is expected to rise from 0.53 billion in 2021 to 1.31 billion by 2050 (1). Patients with T2DM are more susceptible to developing co-morbidities such as cardiovascular disease, renal disease, obesity, stroke, hypertension, and cancer, which are the major contributors to premature death. The treatments currently available for T2DM include lifestyle modifications, injectable insulin, and oral antidiabetic drugs. Although these treatments may demonstrate efficacy for achieving hyperglycaemic control, they are associated with limitations and challenges. For instance, long-term adherence to a controlled diet and exercise requires great self-discipline and persistence; insulin administration requires the precise selection of the injection site, timing, and dosage, and cost considerations should be mentioned as well. The major classes of conventional antidiabetic drugs include α-glucosidase (AG) inhibitors, amylin analogs, biguanides, peroxisome proliferator-activated receptor-γ (PPARγ) agonists, and sodium-glucoseco-transporter-2 (SGLT2) inhibitors (2). Although these agents are relatively more convenient and cost-effective, prolonged administration of these chemical-based small molecules may lead to complications such as gastrointestinal disturbance, nausea, hypoglycemia, edema, and weight gain. These challenges limit the treatments available to individual patients. Hence, considerable efforts have been focused on finding natural antidiabetic therapeutic agents.
Bioactive peptides are defined as peptide fragments, which, upon proteolytic cleavage from the parent protein molecule, exert beneficial bioactivities. These peptides, as a new class of drug candidates, possess several advantages over small molecules used to treat T2DM (3). First, peptides are comprised of amino acids (AA), which are generally more biocompatible and less likely to interfere with normal physiological processes in the host’s body, hence exhibiting fewer side effects compared to synthetic drugs. Second, these peptides can be designed to specifically interact with the targeted enzyme or receptor, bypassing the off-target drawbacks of small drug molecules and concomitantly minimizing the undesired reactions caused by drug-drug interferences when administered with other medications. Third, biomimetic peptides can be engineered to improve their biocompatibility and efficacy in modulating glucose metabolism. For example, Pramlintide, a peptide analog of the glucoregulatory hormone amylin, is currently used in adjunct to insulin in the United States. Fourth, the bioavailability and duration of action of peptides at the target site can be prolonged through various strategies, such as by modifying the peptide’s residues, side chains, terminals, or structure to confer resistance to proteolytic enzymes, obviating the need for frequent dosing, a major drawback of traditional small molecule drugs (4). Overall, peptides offer a promising alternative with unique advantages for treating T2DM. Peptide-based drugs, such as exedin-4, liraglutide, and semaglutide, have already been approved by the US Food and Drug Administration (FDA) during the past decades.
Since the 1990s, targeting AG has been one of the first-line therapeutic strategies for the management of T2DM. As one can imagine, AG inhibitory peptides act by inhibiting the AG enzyme, which is involved in the breakdown of complex carbohydrates into glucose. By inhibiting AG, these peptides can slow down the digestion and absorption of carbohydrates, reducing the rate of glucose release into the bloodstream. This task can be beneficial for individuals with diabetes or those at risk of developing diabetes as it helps regulate blood sugar levels. So, AG inhibitory peptides hold promise as a new class of anti-diabetic agents. By exploring and understanding their properties, mechanisms of action, and structure-activity relationships, this study aimed to identify effective food-based peptides that can be used as potential therapeutics for diabetes. Most AG inhibitory peptides are derived from natural sources such as food proteins. Investigating these peptides provides an opportunity to discover natural and safer alternatives to conventional anti-diabetic drugs. Current anti-diabetic medicines are associated with drawbacks such as adverse side effects and low long-term effectiveness. Therefore, AG inhibitory peptides can broaden our therapeutic options for diabetes, allowing patients to have access to more customized and effective treatments. Studying AG inhibitory peptides allows researchers to acquire a better understanding of the underlying processes involved in carbohydrate digestion and glucose metabolism. This study will advance our scientific understanding of diabetes pathogenesis and our insights into these peptides’ structure-activity relationships and preferences, which may assist in the development of innovative anti-diabetes treatments. Such data can be utilized to develop novel peptide-based pharmaceuticals with enhanced efficacy, selectivity, and bioavailability. The main objective of this study was to explore the potential of AG inhibitory peptides as effective and safe therapeutic agents for the treatment of diabetes and to revisit our understanding of the fundamental mechanisms involved in glucose breakdown regulation.
2. α-Glucosidase Inhibitory Sites
Alpha-glucosidase consists of three main domains: the N-terminal domain (residues 1-346), the C-terminal domain (residues 724-952), and the sub-domain, a (β/α)8 barrel containing the enzyme’s catalytic sites (residues 347-723) (5). The active sites of AG that are involved in the breakdown of glycogen are located deep in the catalytic GH31 domain, which consists of residues Trp376, Trp402, Asp404, Ile441, Asp443, Trp481, Trp516, Asp518, Met519, Phe525, Arg600, Trp613, Asp616, Asp645, Phe649, and His674 (6, 7). Residue Asp282 (from the N-terminal domain) and residue Arg600 are important for substrate identification and stabilization (7, 8), whereas residues Asp518 and Asp616 are the crucial catalytic residues of AG and act as nucleophilic and acid/base catalysts, respectively (5, 9, 10). In patients with T2DM, AG activity is associated with a rise in blood glucose levels secondary to the hydrolysis of polysaccharides and oligosaccharides into monomers (11, 12). Hence, inhibiting the AG active site could delay glucose absorption and maintain postprandial blood glucose at lower levels, reducing the need for insulin (13). Studies have been conducted to explore AG inhibitors, with a focus on bioactive peptides that have a high binding affinity for this enzyme.
In this study, AG inhibitory peptides with low IC50 values (a threshold of 50 mM) were selected from the BioPEP-UMW peptide database (https://biochemia.uwm.edu.pl/biopep-uwm/) (14) and peer-reviewed articles in Scopus (https://www.scopus.com/) (15) to understand their amino acid preferences and their positioning in AG inhibitory peptides. The keyword employed for the literature search in Scopus was “alpha-glucosidase inhibitory peptide”. Inhibitory peptides retrieved were divided into three groups based on the peptide’s length: short (2 - 5 AA residues), medium (6 - 10 AA residues), and long (>10 AA residues). The AG inhibitory peptides explored and their relevant prediction outcomes (i.e., active peptide fragments, binding sites, etc.) were summarized in Appendix 1 (16-32).
The analysis of structure-activity relationship (SAR) on the finally selected inhibitory peptides using Pepsite2 (http://pepsite2.russelllab.org/) (33) showed that Asp518 (96.4%), Asp616 (96.4%), Trp376 (96.4%), Asp404 (89.1%), Ile441 (89.1%), Met519 (89.1%), and Phe649 (85.5%) had the highest binding frequencies. Regarding peptides with different lengths, short inhibitory peptides tend to bind to Trp376 (95%), Asp518 (90%), Asp616 (90%), Met519 (80%), and Phe649 (75%). Also, medium and long peptides showed the same binding frequencies towards the same AG residues: Asp404 (100%), Asp518 (100%), Asp616 (100%), Trp376 (100%), Ile441 (100%), Met519 (90%), His674 (85%), Trp516 (85%), and Phe649 (85%). It could be noted that catalytic sites were of utmost importance for the inhibitory activity of the peptides. Regardless of the peptide length, a high binding frequency was observed toward the enzyme’s catalytic domain, a scenario that was followed by most secondary stabilizer residues. However, the frequency of binding was lower toward substrate-binding sites. Therefore, it was suggested that the inhibitory effects of these peptides were due to the blocking of catalytic sites and the destabilization of AG-substrate complexes.
3. Amino Acids and Their Positions in Inhibitory Peptides
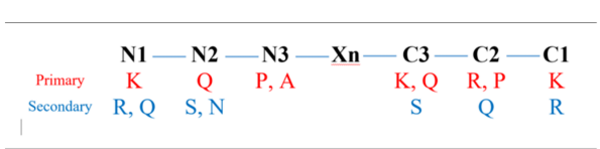
Based on SAR analysis, the types of active AAs and their positioning at the ultimate, penultimate, and antepenultimate structures of both N- and C-terminals were investigated. The positioning could be summarized as follows: N1-N2-N3-Xn-C3-C2-C1, in which N1 was the N-ultimate; N2 was the N-penultimate; N3 was the N-antepenultimate; C1 was the C-ultimate; C2 represented the C-penultimate, and C3 was the c-antepenultimate. The contributions of the AA residue located at these different positions were then predicted by conducting molecular docking using HADDOCK2.4 (https://wenmr.science.uu.nl/haddock2.4/) (34). The X-ray crystallographic structure of human lysosomal AG (PDB ID: 5NN3) was retrieved from the RCSB PDB database (https://www.rcsb.org/) (35). The protein molecule was prepared for docking by removing water molecules and adding hydrogen to the structure using the AutoDock Tools (ADT) program. Following protein preparation, the 3D structures of the peptides were constructed using PepFold3 (bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/) (36-38). The structure with the lowest binding energy was chosen for further analysis of enzyme-peptide interactions using the LigPlot program (39).
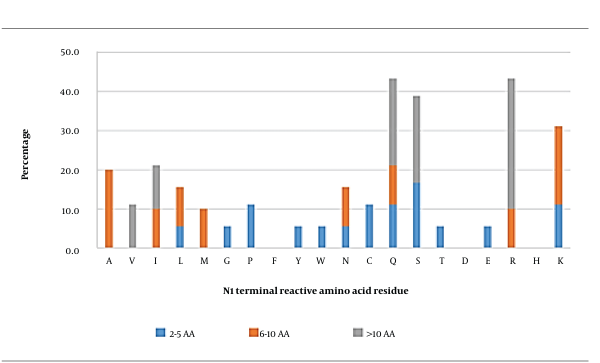
3.1. N-ultimate (N1) Position
In general, glutamine (43.3%), arginine (43.3%), serine (38.9%), and lysine (31.1%) were the most observed reactive AA residues at this position (Figure 1). When the peptides were grouped based on their length, serine seemed to be the most frequently observed reactive AA residue in short peptides. In medium-length peptides, alanine and lysine were the most reactive AA residues, whereas the most frequently observed reactive AA residue in long peptides was arginine. In general, polar neutral or hydrophobic basic AA residues in these peptides were primarily important at this position.
Molecular docking was then conducted for peptides containing glutamine (QITKPN, QQQQQGGSQSQ), serine (SQSPA, SGPFGPK), arginine (RKLKMRQ and RQNIGQNSSPDIYNPQAG), lysine (KLPGF and KLTPQMA), and alanine (ANENIF, AEAGVD) at the N-ultimate position for further analysis (Appendix 2). The results showed that the binding affinity between the peptides and AG was in the range of - 7.0 to - 10.7 kcal/mol. It should be noted that the aforementioned four AAs (glutamine, arginine, serine, and lysine) were highly interactive with AG via hydrophobic bonds as the primary way of interaction. Up to 15 hydrophobic interactions could be observed between KLTPQMA and the residues of Trp376, Asp404, Leu405, Ile441, Trp481, Asp518, Asp616, and Phe649 of AG. Only a trace number of hydrogen bonds and salt bridges were found.
It could be observed that glutamine in QITKPN and QQQQQGGSQSQ formed one hydrogen bond with the Arg411 (2.87 Å) (Appendix 3) and Ser523 (2.90 Å) (Appendix 4) residues of AG, respectively. Appendix 3 and Appendix 4 show that this AA was involved in 7 and 12 hydrophobic interactions, most of which were mediated via an alkyl group. Appendix 5 shows that RKLKMRQ was involved in 4 hydrogen bonds, but none of these bonds were formed by arginine. Similarly, Appendix 6 shows that arginine in RQNIGQNSSPDIYNPQAG formed 2 hydrogen bonds with the Asp282 (2.60 Å) and Asp616 (2.81 Å) residues of AG. In both complexes, this arginine formed 4 and 7 hydrophobic interactions, respectively, which mainly occurred via its alkyl side chain (Appendix 2).
The contribution of the serine residue can be observed from the molecular docking analysis of the peptide sequences of SQSPA and SGPFGPK. Appendix 7 and Appendix 8 show that this amino acid was only involved in 3 and 6 hydrophobic interactions in the SQSPA-AG and SGPFGPK-AG complexes, respectively (Appendix 2). In terms of the lysine residue, Appendix 9 and Appendix 10 revealed that both peptides (KLPGF and KLTPQMA) formed hydrogen bond, each with the residues Asp616 and Asp404, respectively. In addition, this lysine in KLPGF formed hydrogen bond with the residue Asp616 (2.67 Å) of AG (Appendix 9). Meanwhile, three hydrogen bonds were observed between this amino acid in the KLTPQMA sequence and the residues of Asp404 (2.84 Å and 2.94 Å) and Asp518 (2.67 Å) (Appendix 10) of AG. The formation of KLPGF-AG and KLTPQMA-AG complexes also involved 6 and 15 hydrophobic interactions, respectively, which involved the lysine’s alkyl group (Appendix 2). Finally, the contribution of alanine was found to be insignificant. This amino acid in the ANENIF (Appendix 11) and AEAGVD (Appendix 12) sequence contributed to the formation of 2 and 1 hydrophobic interactions, respectively.
It should also be highlighted that the known hotspots (Trp376, Asp404, Ile441, Asp518, Met519, Asp616, and Phe649) were parts of the interactive AG residues. Residues Asp282, Trp481, and Phe525 were also found to frequently form bonds. Therefore, these data supported that these four amino acids could play an essential role in the activity of AG inhibitory peptides, in particular lysine, which could contribute to all types of interactions, followed by arginine or glutamine. These data were supported by Mudgil et al. (10), who stated that the N-terminals of peptides with high affinity for AG were dominated by hydrophobic and basic AAs. The alkyl side chains of lysine and arginine were reported to establish hydrophobic interactions with aromatic residues to enhance the overall stability of the protein-peptide complex (40). Moreover, lysine and arginine are positively charged amino acids, which can be more readily attracted to negatively charged catalytic residues such as Asp518 and Asp616 or stabilizing residues (Asp404, Asp518, and Asp616) in the active site of AG. In this case, the indole side chains of stabilizing residues (i.e., Trp376, Trp516, and Trp613) seem to be potential candidates mediating the cation-π interactions between AG and peptides via lysine and arginine.
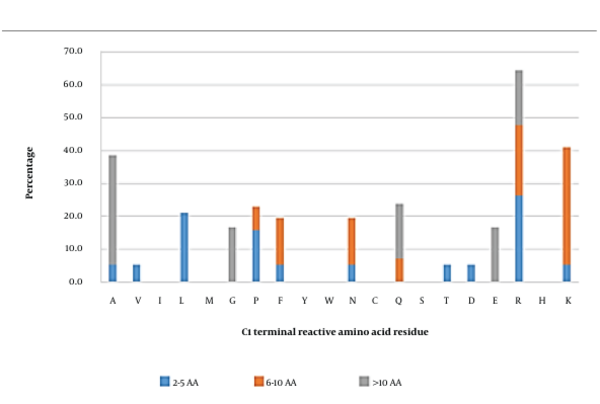
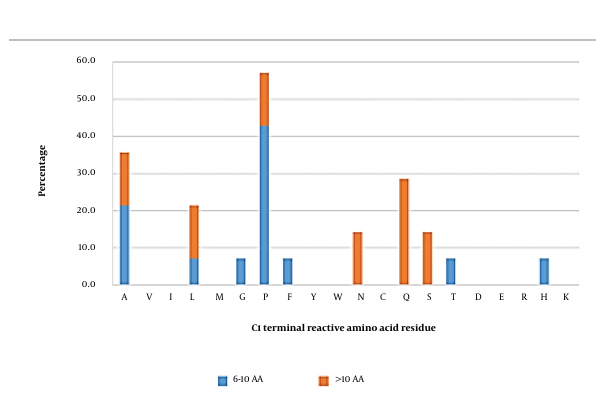
3.2. C-ultimate (C1) Position
Reactive amino acid residues in short inhibitory peptides seemed to be more commonly arginine, followed by leucine (Figure 2). In medium-length peptides, lysine was the most frequently observed reactive residue in the C1 position. On the other hand, alanine was the dominant reactive amino acid in long peptides. Overall, arginine, lysine, and alanine were initially predicted to be significant occupants of the C1 position. Peptides carrying these amino acids were further investigated using docking analysis.
However, docking analysis showed that only arginine and lysine were essentially interacting with AG. Arginine in TPSPR and GSPVSSR formed complexes with AG with the predicted binding affinities of - 8.1 kcal/mol and - 9.7 kcal/mol, respectively (Appendix 2). This amino acid also formed salt bridges with residues Asp518 and Asp616. Appendix 13 and Appendix 14 also showed that this amino acid formed 4 and 3 hydrogen bonds and 16 and 15 hydrophobic interactions with residues Asp518, Asp616, and Leu677, respectively.
Meanwhile, Appendix 15 showed that the side-chain amino group of lysine in GVPMPNK formed 2 salt bridges with the hydroxyl group of Asp616 and 1 hydrogen bond with the residue Asp282 (3.11 Å). Lysine in LLPLPVLK also contributed to 3 salt bridges with Asp518 and Asp616 and one hydrogen bond with Asp518 (2.62 Å) (Appendix 16). In addition, this amino acid in the sequence of GVPMPNK was involved in 8 hydrophobic interactions (Appendix 15), which mainly involved its R group and the Asp616 and Asp282 residues of AG. Appendix 16 showed that lysine in LLPLPVLK contributed to 17 hydrophobic interactions, 16 of which were between its R group and the Arg600, Asp616, Asp518, Trp481, and Met519 residues of AG, as well as one interaction between its hydroxyl group and Leu650 of AG. Alanine, however, only contributed to a few interactions (Appendix 7 and Appendix 17). This could be explained using the similar phenomena mentioned earlier for the N1 position, in which the alkyl side chain formed hydrophobic interactions with the positive charge of their amino groups, forming cation-π interactions with the aromatic residues of AG to enhance the overall stability of the AG-peptide complex (40). Regarding the contribution of 8-16 hydrophobic bonds, 2 - 3 salt bridges, and 1 - 4 hydrogen bonds formed by arginine in TPSPR and GSPVSSR and lysine in GVPMPNK and LLPLPVLK (contributing to 8 - 16 hydrophobic bonds) with the hotspot residues of AG (Trp376, Asp518, Met519, Arg600, Trp613, Asp616, and Phe649), it could be suggested that these amino acids played an essential role at ultimate positions.
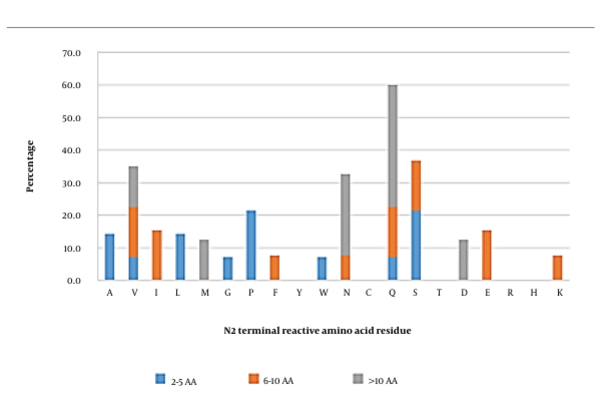
3.3. N-penultimate (N2) Position
In short, peptides, serine, and proline were the amino acids that were able to most frequently interact with AG. Meanwhile, reactive amino acids such as valine, isoleucine, glutamine, serine, and glutamic acid were observed to be the most reactive residues in medium-length peptides. In long peptides, glutamine (37.5%) was found to be the most reactive residue in this position, followed by asparagine (25%). Overall, the residues observed in this position were quite diverse; however, glutamine, serine, valine, and asparagine were the most frequently observed reactive residues, which could interact with AG (Figure 3).
Docking analysis (Appendix 2) showed that only glutamine (in SQSPA and LQAFEPLR) contributed to a significant portion of hydrophobic and hydrogen bonds with AG. Appendix 7 and Appendix 18 show that glutamine in SQSPA could form 2 hydrogen bonds with Asp282 (2.76 Å) and Asp616 (2.97 Å), whereas glutamine in LQAFEPLR was able to form one hydrogen bond with Asp282 (2.61 Å). The ability of glutamine to form multiple hydrogen bonds was attributed to its amide side chain, utilizing 2 lone pairs on the carbonyl oxygen, amine nitrogen, and the 2 hydrogens on the amine group (41). In addition, the glutamine of SQSPA formed 10 hydrophobic interactions (Appendix 2), 9 of which involved the alkyl group and the active site residues of Asp282, Asp616, and Leu650 in AG, as well as one interaction between glutamine’s carboxyl group and the Leu650 residue of AG (Appendix 7). This amino acid also contributed to 9 hydrophobic interactions from the LQAFEPLR-AG complex, 8 of which involved its side chain and the AG residues of Asp282, Leu283, and Asp616, as well as one interaction between the carboxyl group and the Trp481 residue of AG (Appendix 18).
The molecular docking analysis of GVPMPNK and NVLQPS, containing valine at the N2 position, showed that valine was only involved in the formation of the peptide-AG complex via hydrophobic interactions. As shown in Appendix 15, only 2 hydrophobic interactions were observed between the alkyl group of valine in the GVPMPNK peptide and the hydrophobic residues of AG (Leu650 and Ser676). On the other hand, valine in the NVLQPS peptide could form 3 hydrophobic interactions via its alkyl group with the hydrophobic residues of AG (Ala284, Asp616, and Leu650) (Appendix 19).
According to Appendix 20 and Appendix 21, one hydrogen bond was formed between the serine’s hydroxyl group in KSFGSSNI and SSPDIYNPQAGSVT and the Asp282 (2.59 Å) and Lys479 (2.73 Å) residues of AG, respectively. Although seven hydrophobic interactions were also observed between serine in KSFGSSNI and the residues Asp282 and Phe525 of AG (Appendix 20), the other peptide containing serine at the same position did not contribute to any interactions with AG (Appendix 21). A similar trend was found for asparagine at the N-penultimate position (ANENIF, RNPFVFAPTLLTVAAR, and RNLQGENEEEDSGA), as shown in Appendix 11, Appendix 22, and Appendix 23. The only asparagine in ANENIF could mediate binding to AG by forming two hydrogen bonds with Arg600 (2.71 Å) and Asp616 (2.68 Å), as well as via 9 hydrophobic interactions with Asp282, Trp481, Arg600, and Asp616 (Appendix 2). Therefore, arginine seems to have a crucial role at the N2 position, whereas the roles of other amino acids still need to be further investigated. Overall, the hotspots of Asp600 and Asp616 were the interactive sites for this position, along with other residues (e.g., Asp282, Trp481, and Leu650).
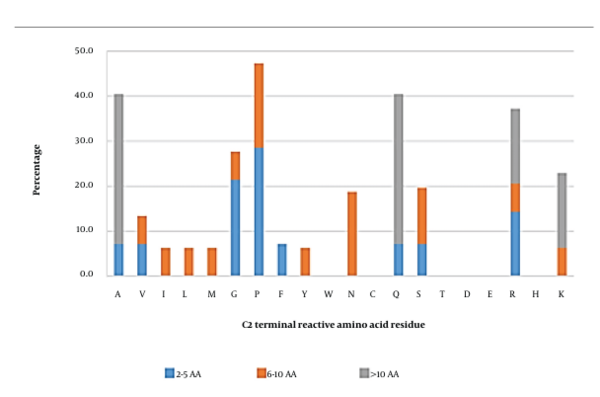
3.4. C-penultimate (C2) Position
The most reactive amino acid at this position was proline (47.3%), followed by alanine, glutamine, and arginine (Figure 4). Short and long peptides were found to have similar reactive amino acids at this position, at which proline, alanine, and glutamine were the most reactive residues, respectively. However, the C2 position in medium-length peptides hosted a variety of reactive amino acids, such as proline (18.8%), asparagine (18.8%), and serine (12.5%). In this scenario, peptides with proline, alanine, glutamine, or arginine at the C2 position were chosen for molecular docking analysis. The results showed that arginine was an essential amino acid (Appendix 24), contributing to the formation of the SWLRL-AG complex by forming 4 salt bridges with Asp518 and Asp616 residues, one hydrogen bond with the Asp518 (2.63 Å) residue, along with 16 hydrophobic interactions, which constituted 51.6% of total hydrophobic interactions within the complex. The RKLKMRQ peptide (Appendix 5) also showed that arginine was involved in the formation of three hydrogen bonds with Asp282 (2.63 Å) and Asp616 (2.59 Å and 3.04 Å), two salt bridges with Asp282, and 6 hydrophobic interactions with Asp282, Asp616, and Leu650.
Proline and glutamine were the next two important amino acids. The docking analysis of SQSPA (Appendix 7) and TPSPR (Appendix 13) showed that proline was involved in the formation of 9 and 5 hydrophobic interactions to generate the SQSPA-AG and TPSPR-AG complexes, respectively (Appendix 2). However, this amino acid did not form any salt bridges or hydrogen bonds for the formation of the SQSPA-AG and TPSPR-AG complexes. The inability of proline to form hydrogen bonds with AG hotspot residues could be due to its relatively rigid side chain (i.e., a five-membered nitrogen-containing ring that binds to the amide nitrogen backbone). The bonding of the amide nitrogen to the side chain would surrender its –NH hydrogen-donating ability. Hence, no hydrogen bond could be formed. Nonetheless, the rigidity of the proline’s side chain may induce steric hindrance at the AG active site, preventing the entry of putative substrate molecules. A similar pattern was observed in the case of glutamine at the C-penultimate position (Appendix 25 and Appendix 26). In the KDLQL (Appendix 25) peptide, glutamine was involved in 9 hydrophobic interactions with AG active site’s residues (Leu678, Leu650, Ser676, Ser679, Leu677, and Trp376), where most of the interactions occurred with the contribution of glutamine’s alkyl side chain, whereas in the SDESTESETEQA peptide (Appendix 26), glutamine at the C-penultimate position formed 3 hydrophobic interactions with the Leu650 and Ser676 of AG.
The positioning of alanine at the C-penultimate site seemed to lead to negligible reactivity, as observed in Appendix 6 (RQNIGQNSSPDIYNPQAG), Appendix 22 (RNPFVFAPTLLTVAAR), and Appendix 27 (VTGRFAGHPAAQ). As shown in Appendix 22 and Appendix 27, alanine at this position could not effectively interact with the active site residues of AG. In the case of the RQNIGQNSSPDIYNPQAG peptide, only 2 hydrophobic interactions were predicted between alanine’s alkyl groups and AG active site’s Phe525 residue. In general, the hotspots of AG (i.e., Trp376, Asp518, Asp519, Asp616, and Phe649) were found to interact with the identified amino acids at the C2 position.
3.5. N-antepenultimate (N3) Position
The most frequently observed reactive amino acid at this position was proline (42.9%), followed by alanine (21.4%), in medium-length peptides, whereas in long peptides, glutamine (28.6%) seemed to be the most frequently observed reactive amino acid (Figure 5). These data suggested that at the N3 position, proline, alanine, and glutamine were most likely reactive residues. Therefore, molecular docking analysis on peptides with various sequences was conducted considering these residues at the N3 position to confirm the interactions involved.
The peptides of GSPVSSR and GFPFYP were chosen to study the involvement of the proline residue at the N-antepenultimate position. The result showed that this amino acid did not form any salt bridge or hydrogen bond with the AG active site (Appendix 2). However, this amino acid interacted with AG via hydrophobic interactions, most of which occurred at its heterocyclic ring (Appendix 14 and Appendix 28). Similarly, molecular docking analysis on the AEAGVD and VVAEQAGEQGFE peptides, which contained alanine at the N-antepenultimate position, revealed that none of the peptides form any salt bridge with AG (Appendix 12 and Appendix 29). One hydrogen bond with a length of 2.82 Å was observed between this targeted amino acid (AEAGVD) and the Arg600 residue of AG (Appendix 12). Moreover, 31 hydrophobic interactions were observed within the AEAGVD-AG complex, 8 of which involved alanine at this position. Meanwhile, 8 out of 50 hydrophobic interactions that occurred within the VVAEQAGEQGFE-AG complex were mediated by alanine at this position (Appendix 29).
On the other hand, glutamine at the N-antepenultimate position of the QQQQQGGSQSQKG peptide (Appendix 30) was not involved in the interactions between the peptide and AG active site, except by only one hydrophobic interaction between the amino group of QQQQQGGSQSQ and a hydrophobic residue (Leu678) of AG (Appendix 4). In conclusion, it can be suggested that proline and alanine at the C3 position could contribute to the stabilization of substrate binding via hydrophobic interactions with AG hotspots (Met519, Arg600, and Asp616). However, glutamine was not important despite the fact that it was frequently located at this position.
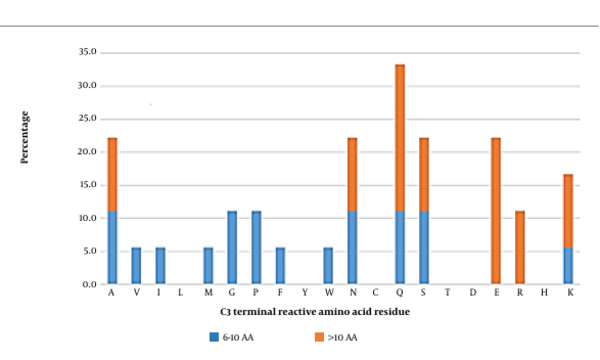
3.6. C-antepenultimate (C3) Position
Glutamine and glutamic acid tend to be the most reactive amino acids at this position in long peptides (Figure 6). Other reactive amino acids towards the AG active site were alanine, asparagine, serine, arginine, and lysine. Medium-length peptides hosted a variety of amino acid residues at this position, suggesting no specific amino acid preferences for the C3 position. However, in general, glutamine, glutamic acid, alanine, asparagine, serine, and lysine seemed to be the most interactive contributors. Further docking analysis was therefore conducted.
Molecular docking analysis on KLTPQMA and QQQQQGGSQSQKG consisted of glutamine at the C-antepenultimate position, which showed that this amino acid was involved in hydrophobic interactions, most of which involved the residue’s side chain (Appendix 10 and Appendix 30, respectively). Glutamine in KLTPQMA and Gln11 in QQQQQGGSQSQKG did not form any salt bridge with the AG active site. However, one hydrogen bond was observed between glutamine in KLTPQMA and the Asp282 residue of AG. Glutamic acid at the C-antepenultimate position in SDESTESETEQA and NALKPDNRIESEGG peptides was observed to only contribute to two hydrophobic interactions towards AG (Appendix 26 and Appendix 31).
The peptides of RNPFVFAPTLLTVAAR (Appendix 22), VTGRFAGHPAAQ (Appendix 27), LAHMIVAGA (Appendix 32), and MIKLRSTAKN (Appendix 33) consisted of a C3 alanine residue. As shown in Appendix 2, this amino acid could not contribute to interactions with AG except in the LAHMIVAGA peptide, where three hydrophobic interactions were observed between the alanine and the Phe525 residue of the AG active site, involving its alkyl groups (Cα and Cβ) and the hydroxyl group.
The data provided in Appendix 11 (ANENIF-AG complex), Appendix 34 (EFLLAGNNK-AG complex), and Appendix 35 (SEDSSEVDIDLGNLG-AG complex) revealed that asparagine at this position did not form any interactions with AG. However, in EFLLAGNNK, Asn7 contributed to one hydrogen bond (length = 2.99 Å) with the Arg281 of AG, as well as one hydrophobic interaction between its hydroxyl groups and the hydrophobic groups of Leu283.
Investigating the potential contribution of serine to the formation of peptide-enzyme complexes suggested that this amino acid could form salt bridges within none of the RNLQGENEEEDSGA-AG (Appendix 23) and YINQMPQKSRE-AG (Appendix 36) complexes. However, this serine formed one hydrogen bond (2.76 Å) with the Arg411 of AG (Appendix 36) and four hydrophobic interactions with Arg411, Trp481, and Phe525 (3 interactions via its hydroxyl groups and one interaction via Cα atom).
The contribution of lysine to the formation of the LAPSLPGKPKPD-AG complex involved 7 hydrophobic interactions via its side chain’s alkyl groups (4 interactions) and amino groups (3 interactions), which interacted with the AG active site’s residues of Asp282, Asn524, and Phe525 (Appendix 37). The lysine residue in QITKPN contributed to the formation of one hydrogen bond with the Asp616 of AG and 8 hydrophobic interactions with the Arg600 and Asp616 of AG (Appendix 2). Overall, glutamine and lysine seemed to have a more pronounced role in forming interactions with AG compared to other amino acids. Similar to the N3 position, amino acids at the C3 position interacted with the hotspots of Met519, Arg600, and Asp616.
4. Perspectives and Conclusions
Peptides suppressing AG activity can be derived from different sources. A variety of these peptides with different sequences can be investigated as potential anti-diabetes agents. This review highlighted the crucial hotspots (i.e., Trp376, Asp404, Ile441, Trp516, Asp518, Met519, Arg600, Trp613, Asp616, Phe649, and His674) of AG involved in interactions with the inhibitory peptides identified. In particular, interactions with the catalytic site of AG were of utmost importance, followed by interactions involved in the stabilization of substrate binding. Yet, the interaction of these peptides with the substrate-binding sites of AG played a less significant role in their inhibitory activity. Therefore, it could be suggested that the inhibitory effects of AG inhibitory peptides were partly due to the blocking of the enzyme’s catalytic sites and the destabilization of the AG-substrate complex. In addition, our data showed that aliphatic alkyl side chains were a key property for the peptides, considering that hydrophobic bonds were essential for their binding to AG. This feature could enhance the binding of the peptide to AG’s hotspots since hydrophobic interactions and π-stacking between the peptide and AG are deemed necessary for the stabilization of the peptide-AG complex and the promotion of inhibitory activity (42). Since the hydrophobic property was of primary importance in this study, it was suggested that the C-H···π-interaction between the peptide and AG played an essential role in their interactions, where residues from both sides could act as either donors or receptors.
Regarding amino acid preferences, as well as the optimized terminal positioning of these amino acids (Figure 7), lysine, glutamine, proline, alanine, and arginine were found to be the primary reactive residues in inhibitory peptides. This can be explained by the fact that lysine is among the top 5 donors in Cα-H···aro-π-interactions (43). Apart from that, lysine is also involved in Cα-H···ac-π-interactions, where it can play a role as a donor at the N1, C1, or C3 position in interactions involving the Asp404, Asp518, and Asp616 (as acceptors) residues of AG. Similarly, alanine is reported to be among the top 2 and top 3 donors in Cα-H···aro-π-interactions and Cα-H···Ac-π-interactions, respectively. Alanine can also be an important player in Cα-H···Arg-π-interactions, Cα-H···Am-π-interactions, and Cali-H···Am-π-interactions. Therefore, it is expected that alanine at the N3 position can significantly contribute to the interaction of an inhibitory peptide with AG. Glutamine is also expected to role as an acceptor (at the N2 or C3 position) and participate in interaction with the Ile441 (as a donor) of AG via Cali-H···am-π-interactions, where the amide group of glutamine can act as a nucleophile (44). In addition, it was reported that the interaction between aromatic residues could be stabilized by a Cα-H···aro-π-interaction involving Pro-Cα-H and the aromatic ring, suggesting that the proline residue (at the C2 or N3 position as a donor) could effectively interact with Trp376, Trp516, and Phe649 (as acceptors) in AG. Finally, arginine at the C2 position facilitates the inhibitory role of the peptide via Cα-H···Arg-π-interactions. Arginine and glutamine could also play their roles at different secondary positions (e.g., N1, C1, or C2). Serine was involved in the same interactions as lysine since both these amino acids are among the top donors in Cα-H···aro-π-interactions and Cα-H···ac-π-interactions. On the other hand, asparagine can be an acceptor when interacting with the hotspots of AG via Cα-H···Arg-π-interactions. In addition, the ultimate terminals (N1 and C1) were found to be highly interactive with the hotspots of AG (Trp376, Asp404, Ile441, Asp518, Met519, Arg600, Trp613, Asp616, and/or Phe649) compared to the penultimate (N2 and C2) and antepenultimate (N3 and C3) terminals. It should also be noted that the effects of neighboring amino acids remain unknown. This review tried to address the features required by researchers for designing or discovering AG inhibitory peptides, taking into consideration their predictive power and accuracy.