1. Background
Antibiotics play a major role in the treatment or prevention of bacterial infections. The earliest use of antibiotics can be traced back to 2500 years ago when moldy soybeans were used by the ancient Chinese to treat furuncle and similar infections (1). In recent years, some of them also have shown anticancer effects (2). For instance, doxorubicin, actinomycin, mitoxantrone, bleomycin, and mitomycin have been registered as antineoplastic therapy agents (3, 4). Although scholars have found many antibacterial active substances in natural plants and animals, microbial antibiotics are still dominant, especially those derived from the Streptomyces genus in soil (5-7). It has been reported that Streptomyces can produce 100,000 antibiotic compounds, accounting for 70 - 80% of all natural bioactive products (8).
Streptomyces can produce a variety of antibiotics, such as streptomycin, tetracycline, biomycin, and actinomycin. With a phenoxazine in its structure, actinomycin D has good anticancer activity (9) and is well-known as an anticancer drug clinically used to treat pediatric tumors and embryonal rhabdomyosarcoma (10). As it can insert deoxyribonucleic acid (DNA) between adjacent guanine-cytosine base pairs to inhibit cell transcription (11), actinomycin D is also an important experimental reagent for determining messenger ribonucleic acid (mRNA) decay rates in cultured cells (12). However, due to its complex chemical structure, fermentation is still the main production of actinomycin D (13), and its low yield limits its wide application.
To expand actinomycin D production, many scholars have made efforts in various ways. Interactions between different species promoted antibiotic production by Streptomyces, such as the addition of paromomycin (14). Some researchers have attempted to isolate new strains to increase actinomycin D production. Djinni et al. (13) discovered a new strain of Streptomyces from Sahara Desert soil and optimized the fermentation of actinomycin D to 656.46 mg/L through the central composite design. Qureshi et al. (15) isolated a novel Streptomyces smyrnaeus from mangrove sediments collected in Saudi Arabia, which simultaneously produced actinomycin X2 and actinomycin D with good antibacterial activity.
In the present study, we isolated and identified potential new actinomycin D-producing strains from the soil, initially optimized fermentation conditions to increase actinomycin D production, purified and identified actinomycin D, and explored the bacteriostatic activity of actinomycin D.
2. Objectives
Based on the current demand for new antibiotic-producing strains, this study specifically screened strains that can produce pigments from purple soil in the Sichuan Basin, purified and identified the pigments produced, and ultimately identified their antibacterial activity.
3. Methods
3.1. Isolation and Identification of Pigment-Producing Bacteria
The strain used in this study was isolated from a soil sample collected from the exposed purple soil in Yanting County, Sichuan Province, People’s Republic of China (PRC). The appearance of the sampling points is shown in Appendix 1, and the information is shown in Table 1.
| Serial Number | Air Temperature | Soil Temperature | Elevation | Coordinates | Slope | Aspect | Land Type |
|---|---|---|---|---|---|---|---|
| LS-2 | 26°C | 19°C | 463 m | 31°15′44″N; 105°24′54″E | 66 ° | Northwest 333° | Exposed rock mass |
The topsoil in the 0~20 cm was collected and packed in an aseptic sealed bag and immediately frozen in the car refrigerator (16). The purple soil extract was prepared and coated on Gause’s synthetic medium (GSM) for culture. The single colony producing yellow pigment was selected and purified 3 times and named LS-2.
The purified strain was sent to Sangon Biotech (Shanghai) Co. Ltd. for sequencing. The determined DNA sequences were compared by BLAST in the National Center for Biotechnology Information (NCBI), and the sequences with high homology were selected to construct the phylogenetic tree in MEGA 7.0 by Neighbor-Joining (NJ) method to determine the taxonomic status of the strain LS-2 (17).
3.2. Physiological Characteristics
The morphology and size of the strains to be identified were observed by scanning electron microscope. The Gram staining method modified by Hucker (18) was used to identify the Gram sex of the identified strains. The mycelium morphology of the strains to be identified was observed by the insertion method (18). The solid plate of GSM was marked parallel and repeatedly, and then the aseptic glass slide was directly inserted into the Agar plate at an angle of 45° and an angle of 90° with the line simultaneously. The carbon source utilization test was carried out according to the method recommended by Berger’s bacterial identification manual (19).
3.3. Purification and Structural Identification of the Yellow Pigment
The bacterial liquid after fermentation was centrifuged under the condition of 8000 r/min for 10 minutes, and then the supernatant was removed by 0.22 μm filter membrane to remove the suspended matter insoluble in water. The vacuum evaporation concentration was under the condition of 100 r/min 60°C, and the concentration was stopped when the amount of fermentation liquid was about 100 mL. The concentrated solution was extracted with ethyl acetate to remove polysaccharides, proteins, and other impurities that may exist in the water. After the extraction, the organic phase was concentrated, the suitable developer was selected on the thin layer chromatographic plate, and the pigment was finally purified by column chromatography (20). The collection solution was concentrated by vacuum evaporation, and the pigment yield was determined by the quality change of the collection bottle before and after. The purified pigment was stored in chromatographic pure methanol, and the purity was verified by high-performance liquid chromatography (HPLC). The structure of the pigment was identified by the ultraviolet-visible (UV-vis) absorption spectrum, IR absorption spectrum, and liquid chromatography-mass spectrometry (LC-MS).
3.4. Optimization of Fermentation Conditions
Five kinds of culture media (21) commonly used for actinomycetes were selected, which were arginine glycerol mineral salt medium (AGMM), glucose nitrate medium (GNM), glycerol-arginine medium (GAM), GSM, and emerson’s medium (EM). The specific formula of the culture medium is shown in Table 2. The strain LS-2 was incubated at a constant temperature of 30°C and 120 r/min in an oscillating incubator for 2 days with an initial pH of 7.0 and 100 mL medium in a 250 mL flask. Under the same conditions, the effect of temperature on the fermentation of strain LS-2 was studied at 26, 30, 34, and 38°C, and the pH was at 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, and 8.5. By comparing the optical density (OD) values of fermentation broth at the maximum absorption wavelength of pigment, the optimal fermentation conditions were determined.
| Medium | Composition (L) |
|---|---|
| Arginine glycerol mineral salt medium | Arginine 1 g, ZnSO4·7H2O 0.001 g, MnSO4·7H2O 0.5 g, MgSO4·7H2O 0.5 g, Fe2(SO4)3·9H2O 0.01 g, CuSO4·5H2O 0.001 g, glycerinum 12.5 g, pH = 7.2 |
| Glucose nitrate medium | K2HPO4·3H2O 0.1 g, NaNO3 0.1 g, KCl 0.1 g, MgSO4·7H2O 0.1 g, pH = 7.0 |
| Glycerol-arginine medium | Glycerinum 20 g, arginine 2.5 g, NaCl 1 g, CaCO3 0.1 g, FeSO4·7H2O 0.1 g, MgSO4·7H2O 0.1 g, pH = 7.2 |
| Gause’s synthetic medium | KNO3 1 g, K2HPO4·3H2O 0.5 g, NaCl 0.5 g, FeSO4·7H2O 0.01 g, MgSO4·7H2O 0.5 g, soluble starch 20 g, pH = 7.2 - 7.4 |
| Emerson’s medium | Beef extract 4 g, peptone 4 g, yeast extract powder 1 g, glucose 10 g, NaCl 2.5 g, pH = 7.0 |
3.5. Determination of Antibacterial Activity
The antimicrobial activity of the extract was evaluated in vitro against a gram-positive bacterium (Staphylococcus aureus) and a gram-negative bacterium (Escherichia coli) by disc diffusion assay (22, 23). The test organisms used in the experiments were reserved by the current research group. Microorganisms were inoculated in lysogeny broth liquid medium (tryptone 10 g/L, yeast extract 5 g/L, NaCl 10 g/L, pH 7.0 - 7.2) and cultured at 140 rpm/min 30°C for 18 - 24 hours. The filter paper discs (6 mm in diameter) were impregnated with the extract and then placed onto the previously inoculated agar plates with the test microorganisms. After the plate was cultured at 30°C for 18 hours, the size of the inhibition zone was measured and recorded to determine the antimicrobial activity.
4. Results
4.1. Physiological Characteristics of LS-2
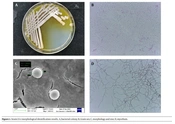
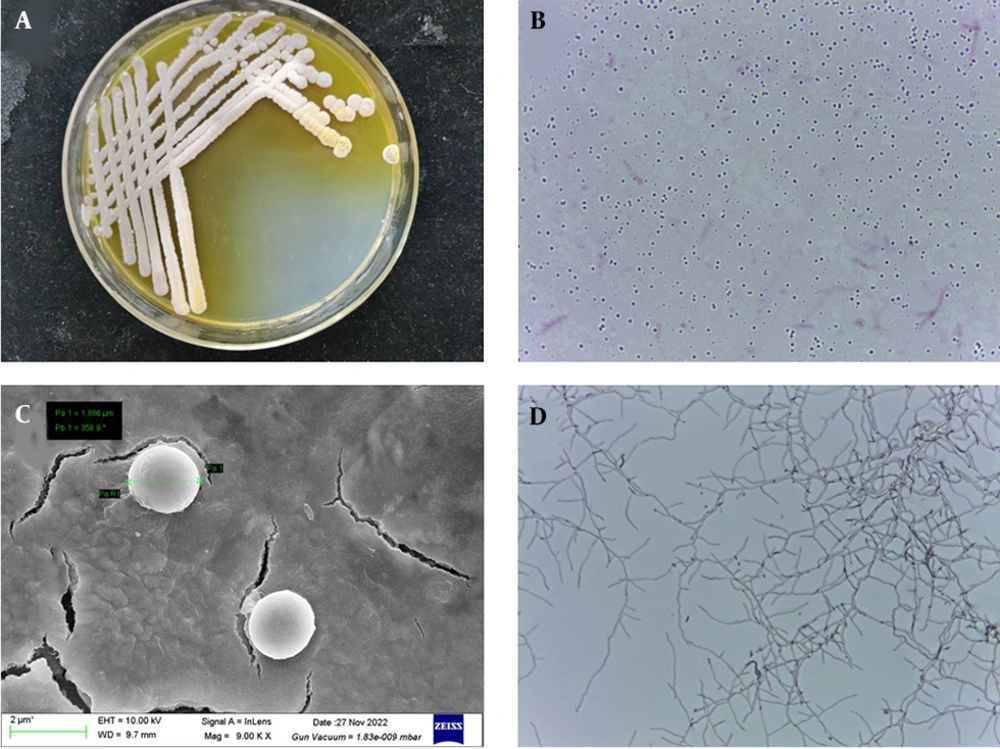
The strain LS-2 formed white and yellowish colored colonies with hydrophilic opaque structure, smooth edges, and irregular dented surface when cultured on Gause’s synthetic agar at 30°C for 2 days. Upon further incubation, the pigmentation of the colony increased and was secreted to the medium (Figure 1A). The strain LS-2 was gram-positive (Figure 1B), the spherical bacterium had a diameter of about 2 μm (Figure 1C). Strain LS-2 had well-developed basal hyphae, aerial hyphae, and spore hyphae, and the hyphae had no diaphragm. The spore filaments are helical in shape, and the number of spirals was mostly 1 circle. At the same time, the spore filaments form conidia, and the spores are spherical (Figure 1D).
The utilization ability of strain LS-2 to different carbon sources is shown in Table 3.
Abbreviations: +, indicates utilized carbon; -, not used; ±, very slight use; /, not detected.
4.2. 16s rDNA Sequence Blast Alignment of LS-2
The sequencing result was compared by BLAST in THE NCBI, and it was found that the homology between LS-2 and Streptomyces parvulus was more than 99%. The sequences with high homology were selected and aligned in MEGA 7.0. The phylogenetic tree was constructed by Bootstrap 1000 times with the NJ method (Appendix 2). The phylogenetic tree demonstrated that the strain LS-2 was on a different node from previously identified strains.
4.3. The Chemical Structure of the Pigment
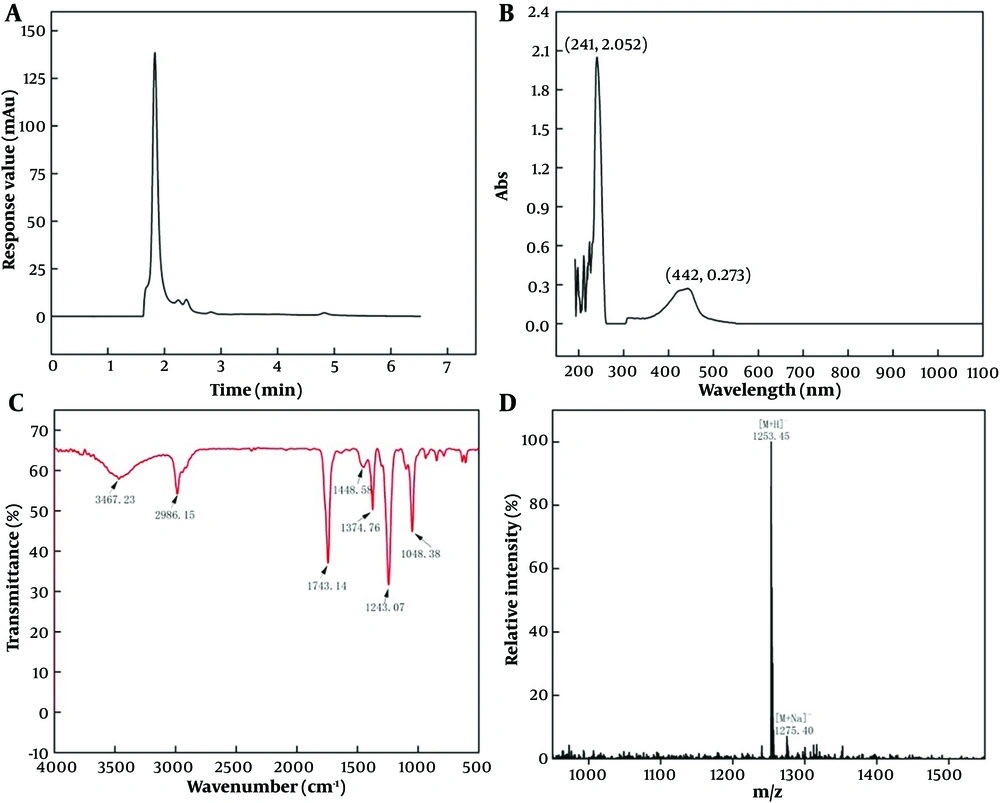
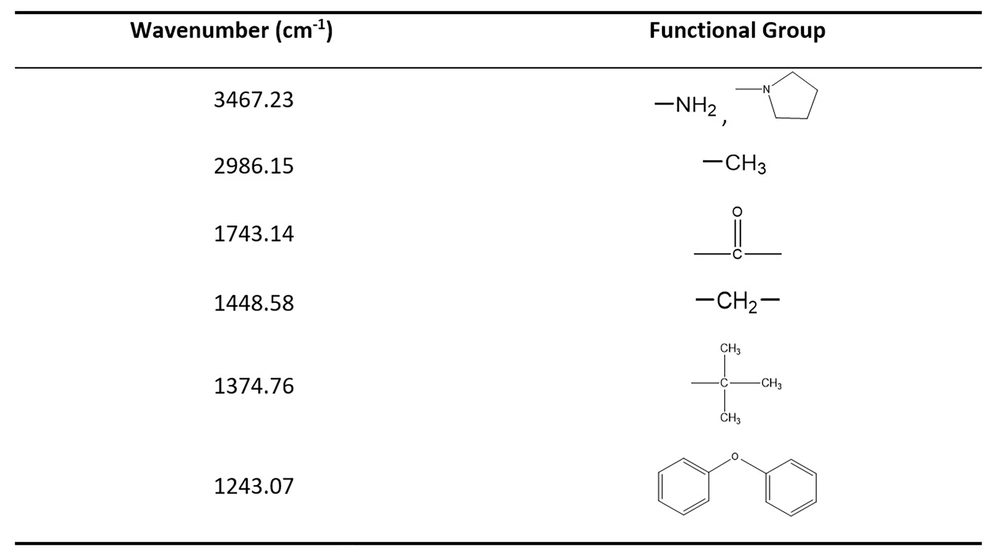
The pigment purification result was verified by HPLC (Figure 2A). The HPLC map showed that the purified pigment had only one main peak, indicating good purity. The obtained pigment has a typical UV-visible absorption spectrum in methanol and has a maximum absorption peak at 221 and 442 nm, which is consistent with actinomycin D (23) (Figure 2B). The results of IR absorption spectra showed that the pigment had absorption peaks at 3467.23, 2986.15, 1743.14, 1448.58, 1374.76, and 1243.07 cm-1. The corresponding functional groups are shown in Figure 3, which accords with the structural characteristics of actinomycin D (Figure 2C). Detected by LC-MS, the pigment had a strongest ion at 1253.45 m/z and a strong ion at 1275.40 m/z, which is the same as actinomycin D (27) (Figure 2D).
Based on the above-mentioned results, the yellow pigment produced by strain LS-2 was identified as actinomycin D, and the structure is shown in Appendix 3 (28).
4.4. The Effects of Fermentation Conditions on Pigment Yield
4.4.1. Effect of Fermentation Medium
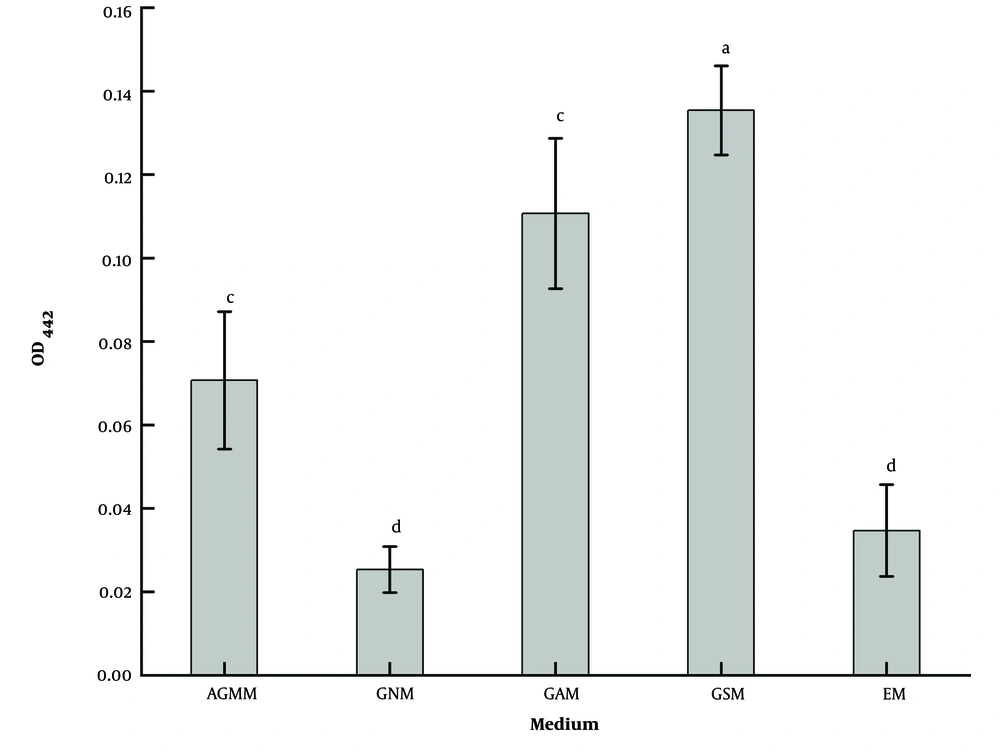
The pigment production ability of strain LS-2 was significantly different on different basic media (Figure 4). The results showed that the strain could grow normally on the common medium of five kinds of actinomycetes; however, the ability to produce pigment was quite different. Gause’s synthetic medium was the best and was significantly better than the other four media (P < 0.05).
4.4.2. Effect of Fermentation Temperature
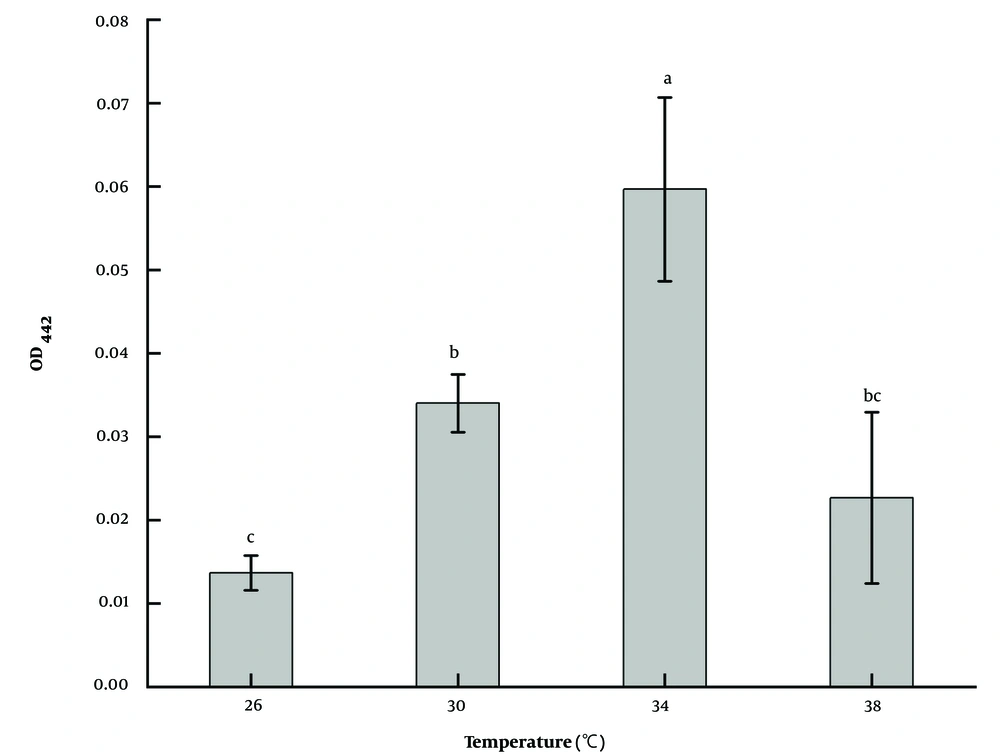
The effect of temperature on the pigment production ability of strain LS-2 was different (Figure 5). The results showed that the strain could grow normally at 26, 30, 34, and 38°C; however, the ability to produce pigment was quite different. After the same culture time, the amount of pigment produced at 34°C was 4.4 times that at 24°C, 1.8 times at 30°C, and 2.6 times at 38°C (P < 0.05). Therefore, 34°C was chosen as the optimal fermentation temperature.
4.4.3. Effect of Culture pH
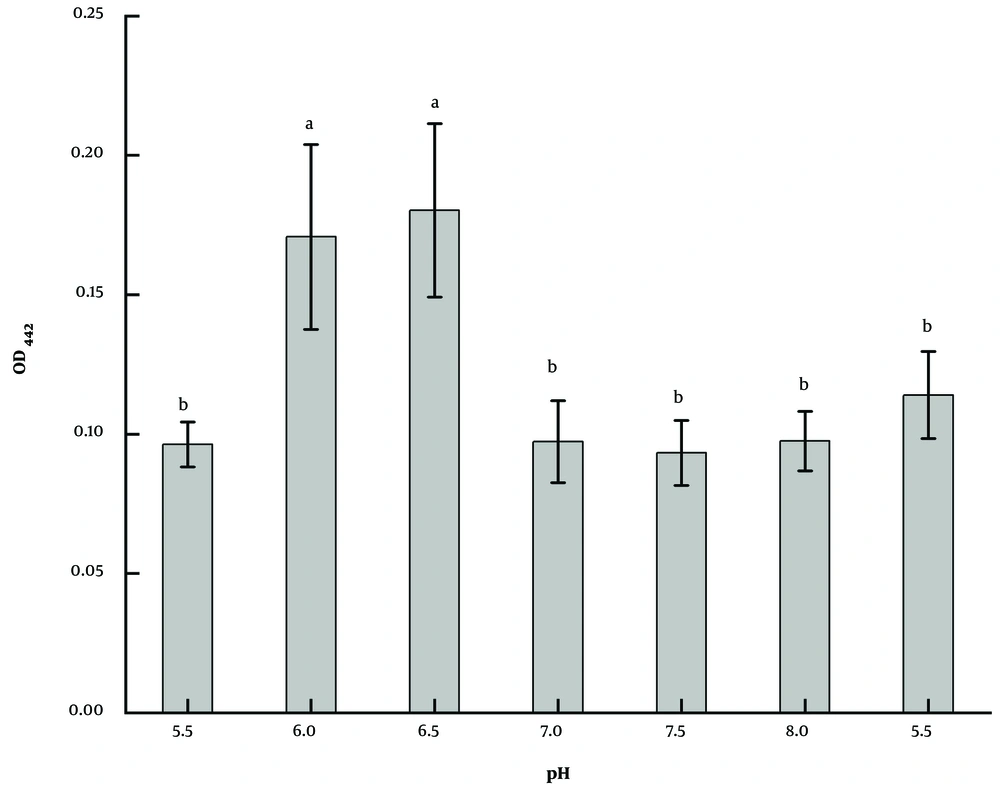
Based on GSM, the strain LS-2 was inoculated after adjusting the medium to a different pH. After 2 days of culture, it was found that the strain could grow normally and had a strong ability to produce pigment in the range of pH 5.5 - 8.5 (Figure 6), among which pH 6.0 and 6.5 were the best, which were about twice as much as those of other groups. The results were significant (P < 0.05). Considering that the medium system is more stable when pH is close to neutral, and pH 6.5 is slightly better than pH 6.0, pH 6.5 is chosen as the optimal initial pH.
4.4.4. Calculation of Pigment Yield
After inoculating strain LS-2 into 6 bottles of 100 mL GSM for 48 hours, 324 mg pigment was finally obtained after filtration, extraction, column chromatography, and vacuum evaporation. All pigments were dissolved in chromatographic pure methanol and stored at 4°C. Therefore, the yield of pigment was 540 mg/L.
4.5. Antibacterial Activity of LS-2
The antibacterial activity of the extract against two pathogenic bacteria was studied by the agar diffusion method. The result is shown in Appendix 4. We found that the extract had antibacterial activity against Staphylococcus aureus and Escherichia coli, and the diameter of the inhibition zone was about 14 mm.
5. Discussion
In this study, we screened a new actinomycin D-producing strain from the Sichuan Basin, preliminarily optimized its fermentation conditions, and identified the structure and antibacterial activity of the actinomycin D produced.
Soil is rich in microbial resources and has many microbial species that have not been observed by human beings. Xie et al. (29) isolated a brown-black pigment-producing strain GXMU-J15T from dry mud and sand and used 16S rRNA gene sequences, phylogenetic tree analysis based on multilocus sequence analysis (MLSA), whole genome sequence construction, and other multiphase methods for taxonomic characterization. The strain GXMU-J15T represented a new species of Streptomyces genus named Streptomyces fuscus sp.nov. Nguyen and Kim (30) isolated a novel actinomycete T113T from forest soil in Pyeongchang County, South Korea, and characterized by multiphase analysis methods, such as 16S rRNA sequence analysis, phylogenetic and DNA-DNA hybridization analysis, chemical genomics, and phenotype analysis. The strain T113T was considered a new species of Streptomyces and named Streptomyces gilvifuscus sp. nov.
We screened a yellow pigment-producing strain LS-2 from purple soil in the Sichuan Basin and identified it by morphology, physiology, biochemistry, and 16s rDNA molecular sequencing. The physiological and biochemical results are shown in Table 2. Compared to other strains producing actinomycin D Streptomyces, it was observed that the characteristics of strain LS-2 were significantly different from those of other actinomycin D-producing strains. The phylogenetic tree also showed that the strain LS-2 was not in the same developmental branch as Streptomyces parvulus. It was speculated that LS-2 might be a new strain.
Optimizing the fermentation conditions of microbial strains is an efficient way to increase the yield of active metabolites. Streptomyces spp. AH11.4 was fermented in tryptone soya broth (TSB) with different carbon sources, and the yield of actinomycin D was 16~305 mg/L (24). Streptomyces flavogriseus NJ-4 was fermented in GS, CS, and GSS medium for 7 days, and actinomycin D reached the highest yield of 960 mg/L in GS (GSM) (26). The fermentation conditions of strain LS-2 were optimized, and the yield of actinomycin D was 540 mg/L after being cultured at 34°C on Gause’s synthesis medium with pH 6.5 for 48 hours. This finding means that a large amount of actinomycin D can be obtained in a short time. It is believed that if the fermentation conditions are further optimized, the yield of actinomycin D will continue to increase.
In addition, this study investigated the antibacterial activity of actinomycin D produced by strain LS-2, and the results showed that it had about 14 mm inhibition zone against both gram-positive bacteria and gram-negative bacteria. Amin et al. (28) screened a Streptomyces from the desert of Egypt and characterized its antibacterial activity with multidrug-resistant Staphylococcus aureus. Moreover, the diameter of the bacteriostatic zone was 4 mm-22 mm. It can be seen that actinomycin D produced by strain LS-2 has excellent and extensive antibacterial activity.
5.1. Conclusions
In this study, we screened an actinomycin D-producing strain of Streptomyces parvulus LS-2 from the Sichuan Basin. Through the comparison of physiological and biochemical characteristics and phylogeny, it was found that it is a potential new strain. After the preliminary optimization of its fermentation conditions, LS-2 showed a high yield. Using Staphylococcus aureus and Escherichia coli as the reference of gram-positive and gram-negative bacteria, respectively, the actinomycin D produced by strain LS-2 showed excellent antibacterial activity.