1. Background
The herb Salviamiltiorrhiza Bunge (Lamiaceae) is widely recognized in traditional Chinese medicine for its exceptional efficacy in ameliorating microcirculatory disturbances (1). Tanshinones, a group of terpenoids comprising tanshinone I, dihydrotanshinone I, tanshinone IIA, and cryptotanshinone, represent the primary bioactive constituents found in S. miltiorrhiza. Tanshinone IIA is commercially synthesized from the dried roots of S. miltiorrhiza, a process commonly employed in the industrial production of this compound. In recent years, various biological benefits of tanshinone IIA have been discovered, including the treatment of myocardial infarction, coronary heart disease, and angina (2-4). It also demonstrates substantial anti-tumor, antiviral, and neuroinflammatory alleviating effects (5-8). Due to excessive collection of wild resources, the natural supply of this medicinal plant is currently limited. Various techniques have been employed to enhance tanshinone production from S. miltiorrhiza roots, including tissue cultures, microwave-assisted extraction (MAE) (9), beta-aminobutyric acid induction (10), and biotic and abiotic elicitors (11); however, the issue of resource scarcity for S. miltiorrhiza remains unresolved.
The potential of endophytes as reservoirs of natural products for industrial, agricultural, and pharmaceutical applications has been widely recognized in recent decades, particularly regarding the secondary metabolites produced by fungal endophytes inhabiting medicinal plants (12). However, the current production of highly potent secondary metabolites reported in various endophytes, such as Gentiana macrophylla (13), Icacinaceae (14), and Forsythia suspensa (15), remains limited. The endophytic fungus Emericella foeniculicola TR21 (Aspergillus) was isolated from the roots of S. miltiorrhiza to explore an alternative natural source of tanshinone IIA production. However, the yield of tanshinone IIA (YT) also remained relatively low (16).
The protoplast mutation is a highly effective approach for addressing this issue within different mutation breeding techniques, resulting in the acquisition of improved mutants through mutagenic or chemostat-mediated adaptation. In this process, the cytomembrane of isolated protoplasts is fully exposed, rendering them highly susceptible to the external environment. Previous studies have reported that Cordyceps militaris SU5-08 was obtained by subjecting the initial strain (C. militaris SU5) to ultraviolet mutagenesis of protoplasts, resulting in an extraction rate of exopolysaccharide (EPS) at 1919.16 ± 165.27 mg/L, which surpassed that of C. militaris SU5 (17). The marine-derived fungus Aspergillus sp. NRCF5 was utilized to induce genetic variability through the application of various combinations and doses of mutagens. Five mutants exhibiting high xylanase activity and a tolerance of 0.25% (w/v) to the antimetabolite 2-deoxyglucose (2DG) were isolated from populations generated by combining UV and NTG mutations (18). The protoplast mutation method can thus be employed for TR21 to obtain mutant strains with enhanced tanshinone IIA yield, establishing a novel pathway for the production of tanshinone IIA.
2. Objectives
The objective of this study was to select a mutant with enhanced tanshinone IIA production through protoplast mutation induced by UV and Sodium nitrite (NaNO2), thereby offering an alternative and high-yield natural source of tanshinone IIA. This establishes a cost-effective and non-plant-derived substitute for tanshinone IIA.
3. Methods
3.1. Reagents and Instruments
Snailase, lysozyme, and cellulase were obtained from Sigma Chemical Co., Ltd. (Nanjing, China). Sodium nitrite was acquired from Hongyan Chemical Reagent Co., Ltd (Tianjin, China). Tanshinone IIA was purchased from the Shaanxi Provincial Branch of the State Medical Products Administration (Shaanxi, China). Chromatography-grade methanol was procured from Tianjin Kermel Chemical Reagent Co., Ltd (Tianjin, China). All other reagents were of analytical grade and sourced from commercial suppliers unless otherwise specified.
Scanning electron microscope (SEM) micrographs were captured using a S-570 SEM (Hitachi, Japan) at 15 kV. A ZF-2 ultraviolet lamp (Shanghai Anting Electronic Instrument Factory, China) was used for UV irradiation. Mycelium pellets were disrupted using the BILON88-II ultrasonic cell crusher (Shanghai Bilang Instrument Co., Ltd., China). The initial YT was detected using a TU-1810 UV/Vis Spectrophotometer (Beijing Puxi General Instrument Co., Ltd., China). The production of tanshinone IIA was accurately determined using an LC-2010A high-performance liquid chromatography (HPLC) (Shimadzu, Japan).
3.2. Strains and Culture Medium
The fungus E. foeniculicola TR21 (Accession No. HQ840704) was isolated from the roots of S. miltiorrhiza (16) and cultivated on potato dextrose agar (PDA) medium at 28°C for 7 days. The culture was then stored at 4°C in the Laboratory of Applied Microbiology, School of Life Sciences, Shaanxi Normal University.
The PDA medium consisted of diced potatoes (200 g/L), dextrose (20 g/L), and agar (20 g/L), with a natural pH. The composition of potato dextrose broth (PDB) medium is identical to that of PDA, excluding the presence of agar. Phosphate buffered saline (PBS) used for protoplast formation was composed of Na2HPO4·12H2O (0.2 M), NaH2PO4 (0.2 M), and NaCl (0.8 M), with a pH of 6.0. The protoplast regeneration medium (PRM) was prepared by supplementing PDA medium with PBS.
3.3. Protoplast Mutation and Mutant Screening
The spores of TR21 were suspended in 20 mL of PBS and gently shaken for 20 minutes at room temperature. The treated spores were subsequently harvested through centrifugation at 3000 rpm for 10 minutes, followed by resuspension and incubation for 3 hours at 28°C in lytic enzyme solutions containing snailase (2%), lysozyme (1.5%), and cellulase (1.5%). The protoplasts were then purified via filtration using a small mass of cotton and collected through centrifugation at 2000 rpm for 10 minutes. The resulting pellets were then subjected to two washes with PBS.
The purified protoplast suspension was exposed to UV radiation (15 W, 30 cm above the suspension) for a gradient of six durations (1.5, 2.0, 2.5, 3.0, 3.5 and 4 minutes) in the absence of light, followed by treatment with NaNO2 at a concentration of 1.5 g/L. The protoplast suspension was then diluted in a gradient manner 104 times with PBS, and 0.1 mL of the diluted solution was spread on the PRM plate. The initial protoplast suspension of TR21 was used as a control. After incubation for 7 days at 28°C, the larger colonies were selected and transferred onto new plates. Subsequent rounds of mutation were conducted through protoplast mutation induced by UV and NaNO2.
3.4. Extraction of Cell-associated Tanshinone IIA
The TR21 and mutant strains were cultured in 50 mL of PDB at 28°C for 7 days. Subsequently, the resulting fermentation broths were centrifuged at 5000 rpm for 30 minutes to obtain mycelium pellets. These pellets were then washed with sterile distilled water, dried at 70°C until a constant weight was achieved, powdered using a glass mortar and pestle, and extracted with 6 mL of methanol using a BILON88-II ultrasonic cell crusher. All samples were filtered twice through disposable syringe filtering cartridges equipped with 0.22 μm nylon membranes before being subjected to preliminary detection via a TU-1810 ultraviolet-visible (UV/Vis) spectrophotometer. Samples obtained from improved mutant strains underwent more accurate detection using an LC-2010A HPLC.
3.5. Preliminary Detection by Ultraviolet-Visible Spectrophotometer
The samples were diluted to specific percentages and subsequently analyzed using quartz cuvettes (5×1 cm) in a TU-1810 UV/Vis spectrophotometer at a wavelength of 270 nm. Methanol was used as the control in the experiment. Absorption measurements were performed three times to obtain averaged absorption data.
The regression equation was determined as follows: OD270 = 0.091C + 0.0631, R² = 0.9994 (N = 6), where OD270 represents the absorption of tanshinone IIA at a wavelength of 270 nm, and C denotes the concentration of tanshinone IIA (mg/L). The linearity of the detector response within the range of 1.9392 to 7.272 mg/L enabled accurate quantification.
3.6. Accurate Detection by High-Performance Liquid Chromatography
The HPLC analysis was conducted using an Inertsil ODS chromatographic column (5 μm×4.6 mm × 250 mm) from Dikma, maintained at 30°C with the assistance of a column heater. After performing ultraviolet absorption measurements on tanshinone IIA, the optimized absorption wavelength of 270 nm was selected. Prior to use, the mobile phase, consisting of methanol and water (85:15, v/v), was filtered and degassed through a PTFE filter (4 mm × 0.45 mm). The flow rate was set at 0.8 mL/min for each run, with a total run time of 40 minutes per injected sample. To obtain reliable results, three injections were made for each sample with an injection volume of 30 μL.
The regression equation was determined as follows: A = 6.22 × 107 C - 33147, R² = 0.9994 (N = 5), where A represents the peak area of tanshinone IIA and C denotes the concentration of tanshinone IIA (mg/mL). The detector response exhibited linearity within the range of 0.0064 to 0.064 mg/mL, enabling accurate quantification using this method, which demonstrates high sensitivity and accuracy.
3.7. Stability Assessment of Mutant Strains
The selected highest-yielding strains were subjected to continuous cultivation, and the tanshinone IIA content was determined using the TU-1810 UV/Vis spectrophotometer with three samples for each generation. After five generations, the dry weight (DW) and YT in the first generation were used as controls to assess the passage stability of the high-yield mutant strains.
3.8. The Genetic Change Between the Wild-Type and the Mutant
The fungal genomic DNA of both wild-type and mutant strains was isolated from axenic cultures using a modified cetyltrimethyl ammonium bromide (CTAB) protocol (19). The fungal mycelia were meticulously harvested from fresh cultures growing on PDA plates at 28°C for 7 days using sterile nippers and then suspended in a solution containing 120 μL of acid sodium dodecyl, 30 μL of β-Mercaptoethanol, and 600 μL of CTAB buffer. The subsequent steps of the extraction procedure were conducted according to the instructions outlined in the Kit User's Manual. The extracted DNA pellet was subsequently preserved in 50 μL of tris-EDTA (TE) buffer at -20°C.
The genetic changes between the wild-type and mutant strains were assessed using 20 randomly selected 10-mer primers (Sangon Biotech Co., Ltd., Shanghai, China). Amplification reactions were carried out in a 25 μL mixture containing 0.4 μL of 10 × Taq DNA polymerase buffer (including Mg2+), 0.5 μL of dNTPs (10 mM), 1.2 μL of each primer (10 μM), 0.4 μL of Taq polymerase, and 1 μL of fungal genomic DNA. The thermal cycling procedure was conducted as follows: An initial denaturation at 94°C for 5 minutes, followed by 40 cycles of denaturation at 94°C for 60 seconds, primer annealing at 36°C for 90 seconds, extension at 72°C for 60 seconds, and a final extension at 72°C for 10 minutes. The gels were stained with ethidium bromide (EB) and visualized under ultraviolet light.
3.9. Statistical Analysis
The experiment was conducted with three replicates for each treatment, and the results are presented as mean ± standard deviation (SD). The data were analyzed using SPSS 20.0. Statistical differences between the treatment and control groups were assessed using an independent-samples t-test. The figures indicated significant differences compared to the control group, with * P < 0.05 and ** P < 0.01.
4. Results
4.1. Protoplast Mutation and Mutant Screening
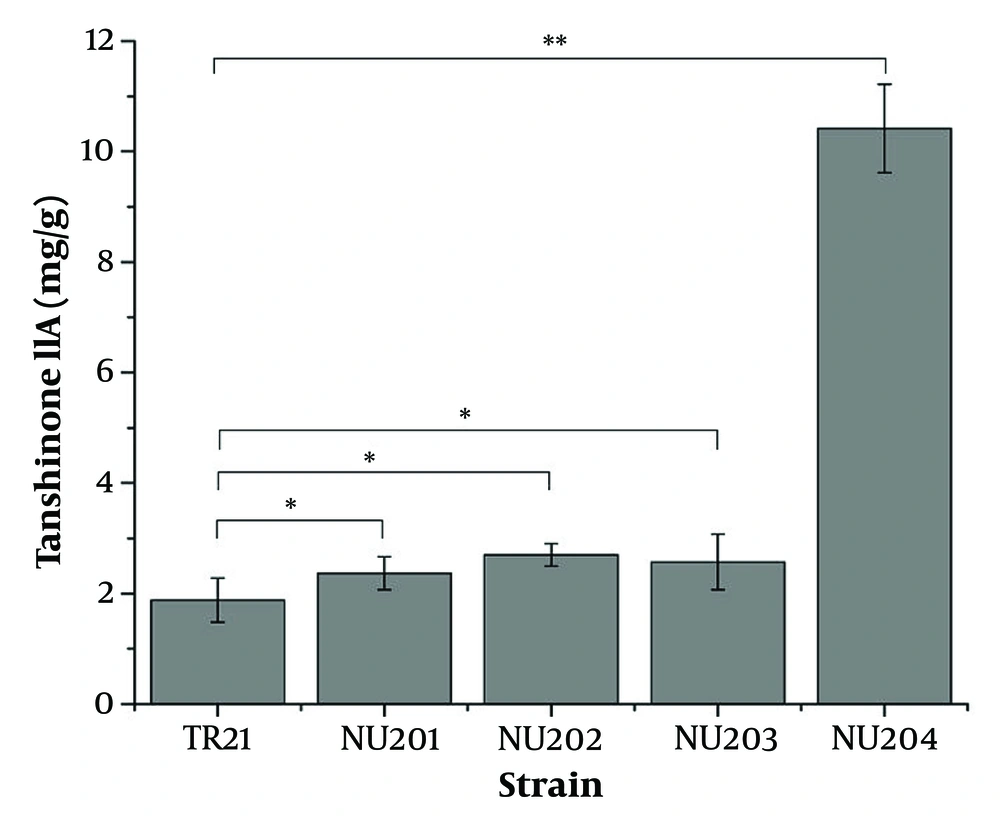
The enhanced production of tanshinone IIA through protoplast mutation induced by UV and NaNO2 is illustrated in Figure 1. Among the three hundred colonies grown on PDA medium, four were selected for their significantly higher tanshinone IIA yield compared to the wild-type TR21 (1.89 mg/g), as determined by TU-1810 UV/Vis spectrophotometer. The production levels of tanshinone IIA were significantly higher in the four isolates (NU201, NU202, NU203, and NU204), with values of 2.37, 2.70, 2.57, and 10.42 mg/g, respectively. The highest-yielding mutant strain was NU204, which demonstrated a production capacity of tanshinone IIA 5.51 times greater than that of TR21 (P < 0.01).
Improved production of tanshinone IIA by protoplast mutation of ultraviolet radiation (UV) and sodium nitrite (NaNO2). Four out of three hundred colonies were examined for a higher yield of tanshinone IIA (YT) than the wild-type TR21 by TU-1810 ultraviolet-visible (UV/Vis) spectrophotometer. * P < 0.05, ** P < 0.01, where comparison between the mutant and the wild-type, was presented in the figure. Error bars represent standard deviation (SD) of three independent experiments.
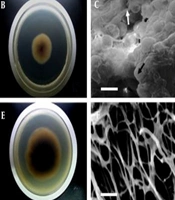
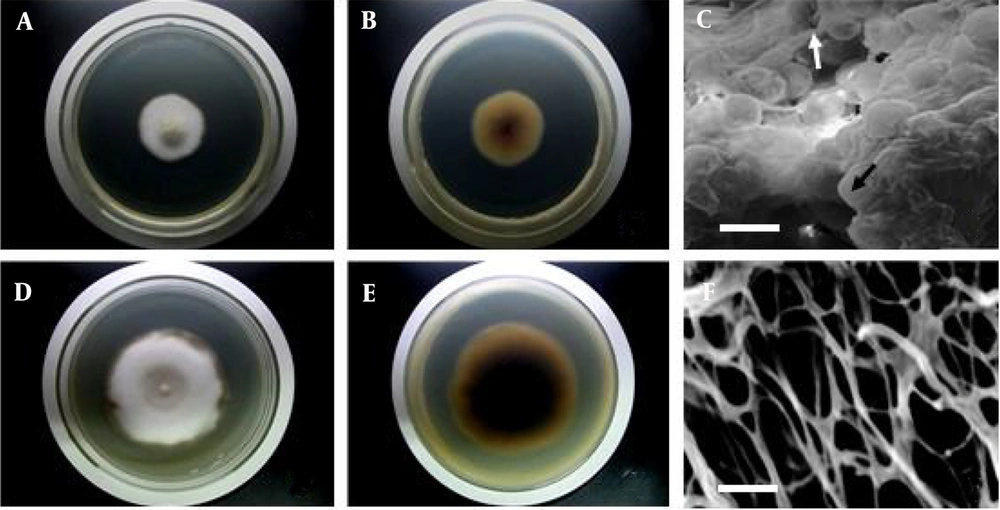
In addition to alterations in the production of tanshinone IIA, there were also significant variations in the colony morphological characteristics of NU204. The changes in colony morphology are depicted in Figure 2. After 7 days of cultivation at 28°C on PDA medium, the colonies of NU204 exhibited a larger size compared to TR21 (Figure 2A, D). Furthermore, the underside of NU204 colonies appeared darker than that of TR21 (Figure 2B, E). Notably, cross-linking was observed between the endophytic hyphae and perithecium of TR21 (Figure 2C). The hyphae of NU204 are depicted in Figure 2F, while the presence of perithecium was not observed.
Colony morphological characteristics of TR21 and NU204. (A) and (D) show the front of TR21 and NU204 on PDA after 7 d, respectively. (B) and (E) show the back of TR21 and NU204 on PDA after 7 d, respectively. (C) shows the scanning electron microscope (SEM) micrograph of aggregated mycelium (white arrow) and round cleistothecium (black arrow) of TR21. (F) shows the SEM micrograph of aggregated mycelium of NU204. Scale Bar = 30 μm.
4.2. The Production of Cell-Associated Tanshinone IIA from NU204
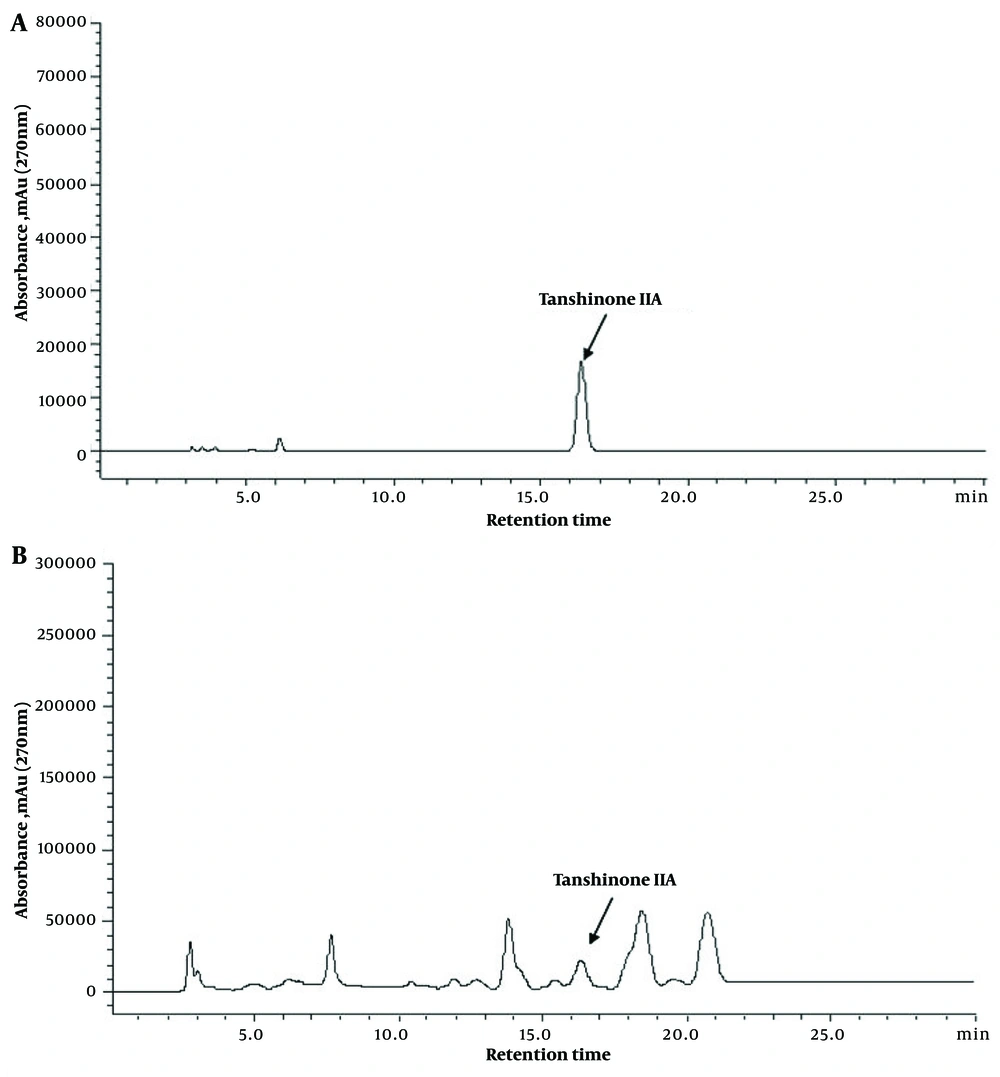
The HPLC results revealed that the authentic tanshinone IIA had a retention time of 16.357 minutes (Figure 3A). NU204 exhibited a relatively high YT at the same retention time, with a value of 165 ± 2.52 μg/g (Figure 3B). Conversely, TR21 demonstrated a comparatively low yield for tanshinone IIA production (16). The tanshinone IIA yield in the mutant NU204 was significantly increased by 4.67-fold compared to the wild-type TR21 (P < 0.01), indicating a significant enhancement in NU204 production compared to TR21.
4.3. The Genetic Stability of NU204
The highest-yield mutant strain NU204 was subjected to continuous cultivation for five generations, during which the DW and YT were assessed for each generation, with NU204-1 serving as a control. The results revealed that all generations demonstrated comparable DW and YT values (Table 1). There were no significant differences in DW and YT among the different generations (P > 0.05), indicating the genetic stability of NU204 and its practical applicability.
| Generation | DW (g/50 mL) | YT (mg/g) |
|---|---|---|
| NU204-1 | 0.34 ± 0.02 | 10.28 ± 0.12 |
| NU204-2 | 0.33 ± 0.02 | 10.46 ± 0.11 |
| NU204-3 | 0.31 ± 0.02 | 10.54 ± 0.14 |
| NU204-4 | 0.30 ± 0.01 | 10.39 ± 0.11 |
| NU204-5 | 0.30 ± 0.01 | 10.21 ± 0.12 |
a DW is dried weigh of mycelium; YT is the yield of tanshinone IIA.
4.4. The Genetic Change Between TR21 and NU204
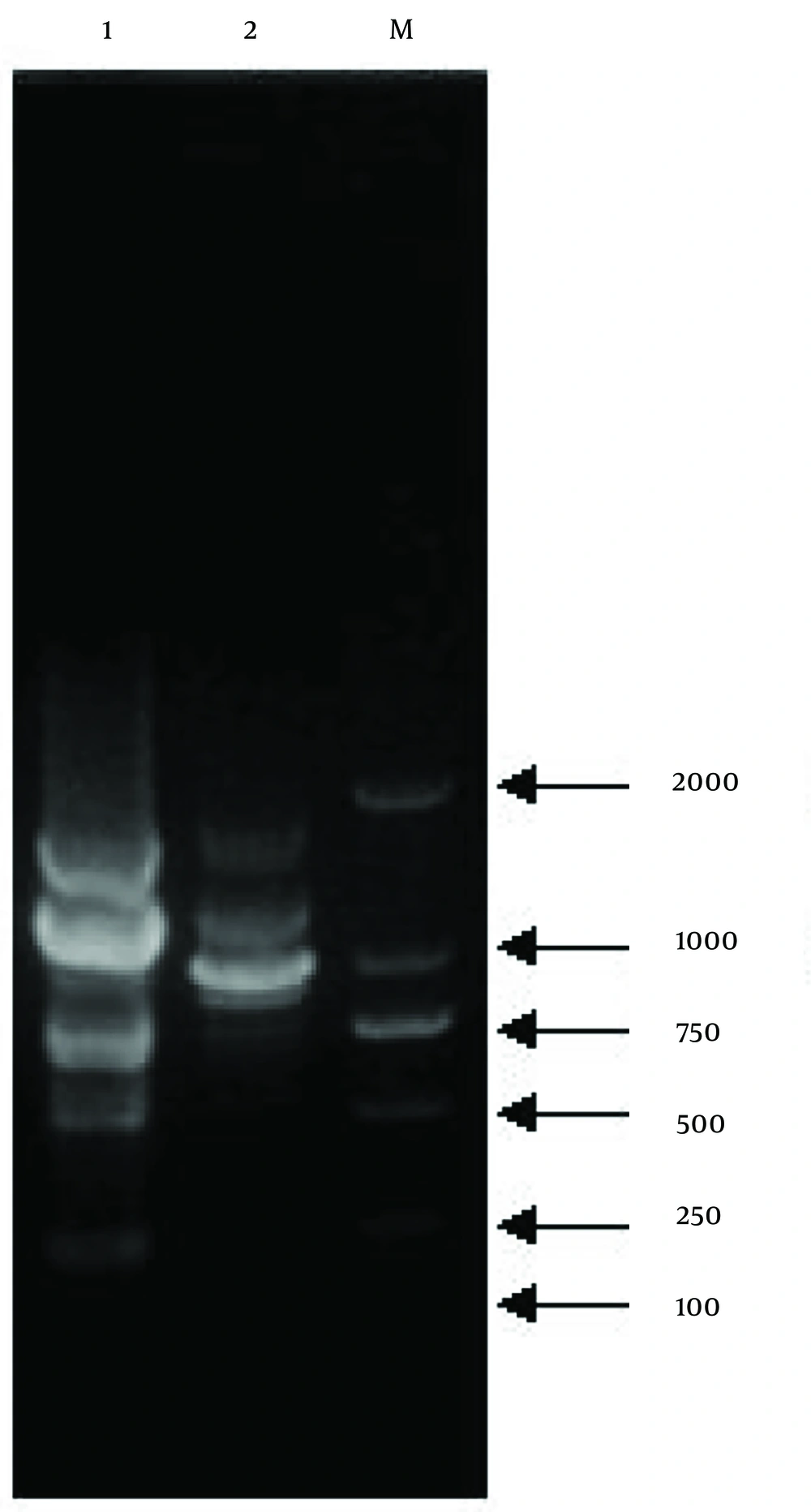
The TR21 and NU204 strains were subjected to analysis using 20 randomly selected 10-mer primers (S61 - S80). The S75 primer (5'-GACGGATCAG-3') generated polymorphic bands, and the amplified fragments were characterized based on their size, which ranged from approximately 100 - 2000 kb (Figure 4). The observed disparity in molecular weights of DNA fragments between TR21 and NU204 suggested a potential genetic alteration induced by protoplast mutation, indicating the presence of distinct genetic variations.
5. Discussion
The production of tanshinone IIA is enhanced through protoplast mutation induced by UV and NaNO2. In this process, the cytomembrane of isolated protoplasts is fully exposed, making them highly susceptible to the external environment. The primary mechanism underlying UV-induced mutants can be attributed to the formation of thymine dimers, which alter the DNA structure and ultimately lead to microbial variation (20, 21). Sodium nitrite induces mutants likely through a different mechanism, where the deamination of purine or pyrimidine in DNA by NaNO2 leads to alterations in the structure and properties of nucleic acids, ultimately disrupting DNA replication. The combined effects of UV and NaNO2 can be achieved through multi-site alterations, reducing the reverse mutation ability and stabilizing genetic variations in traits. This is accomplished by combining a narrow range of physical mutations with varying intensities of chemical mutagenesis (22).
To assess the genetic changes between TR21 and NU204, a set of 20 randomly selected 10-mer primers was utilized to amplify genomic DNA from both strains. The results strongly indicate that the observed genetic alterations in TR21 and NU204 can be attributed to protoplast mutation. Notably, multiple modifications have occurred at various sites within the DNA sequences of these two strains. Furthermore, alternative breeding methods could also be explored to generate more desirable mutants (23, 24).
The ability of TR21 and NU204 to synthesize tanshinone IIA supports the theory that, during the long co-evolution between endophytes and their host plants, endophytes adapt to their specific microenvironment through genetic variation, including the uptake of some plant DNA into their genomes (25). The potential of utilizing endophytes as novel sources of natural products in medicine, agriculture, and industry is highly promising. Research studies have focused on the gene expression of S. miltiorrhiza, leading to tanshinone synthesis. By successfully isolating tanshinone biosynthesis genes, such as SmMDS, SmDXS, SmDXR, SmHMGR, and SmGGPPS, from S. miltiorrhiza, attempts have been made to enhance the production of tanshinone in S. miltiorrhiza through genetic engineering (26-29). The mevalonate (MVA) pathway and the methylerythritol phosphate (MEP) pathway have been identified as key pathways in tanshinone biosynthesis in S. miltiorrhiza. The MVA pathway primarily contributes to cell growth, while the MEP pathway supplies precursors for diterpenoid biosynthesis (30). Therefore, it is possible that analogous tanshinone biosynthesis genes and pathways may be present in tanshinone IIA-producing fungi. Further investigation should prioritize elucidating the regulatory mechanisms underlying tanshinone IIA biosynthesis.
The wild-type TR21 was isolated from the roots of S. miltiorrhiza to explore an alternative natural source for tanshinone IIA production. However, the YT produced by TR21 is relatively insufficient to meet the requirements for industrial-scale production (16). The protoplast mutation method employed in this study proves valuable for rapid breeding, ultimately resulting in the development of the genetically stable mutant NU204 with enhanced tanshinone IIA production. The production of tanshinone IIA in NU204 was significantly increased by 4.67-fold compared to TR21, indicating a significant enhancement in NU204 yield relative to TR21. The mutant NU204 is currently emerging as a promising novel alternative source for tanshinone IIA, offering potential practical applications in production.