1. Background
The escalating issue of antimicrobial resistance underscores the urgent need for alternative therapeutic strategies, with probiotics emerging as a promising option for addressing various diseases. Probiotics present a valuable approach to combating pathogenic colonization and enhancing host defenses (1, 2). The primary bacterial genera utilized as probiotics include Lactobacillus, Lactococcus, Staphylococcus, Enterococcus, Pediococcus, Bifidobacterium, Propionibacterium, Bacillus, and Escherichia (3). Although probiotics are not a direct replacement for antibiotics, they exert antimicrobial effects through multiple mechanisms. One such mechanism involves competition for resources and adhesion sites. Probiotics colonize the gut, thereby restricting the space and nutrients available to pathogenic bacteria. Additionally, certain probiotics produce bacteriocins—protein-based molecules that specifically target and kill pathogens (4-6).
Probiotics can also modulate the host immune response, strengthening defenses against invading bacteria (7). When used strategically, specific probiotics may enhance treatment outcomes for infections caused by resistant bacteria. Probiotics produce various inhibitory compounds, including organic acids, hydrogen peroxide, proteinaceous substances like bacteriocins and antibacterial peptides, and other antimicrobial agents (8, 9).
Antimicrobial peptides and bacteriocins present in the cell-free supernatants (CFS) of probiotics, particularly lactic acid bacteria (LAB), have demonstrated the ability to inhibit the growth of a broad spectrum of pathogenic bacteria, fungi, and viruses. Additionally, these small peptides and proteins contribute to stabilizing gut barrier function (10).
The intricate interactions between microbial communities residing in the human body and their impact on health have become a focal point in biomedical research. Staphylococcus epidermidis, a common member of the skin and mucosal microbiota, has emerged as a significant opportunistic pathogen, particularly in the context of urinary tract infections (UTIs) among hospitalized patients (11, 12).
Despite its predominantly commensal role, S. epidermidis can exploit host vulnerabilities to cause infections, often linked to biofilm formation and resistance to conventional antibiotics. While S. epidermidis is rarely associated with UTIs, its prevalence in urine samples has been reported to range from 42.3% to 62.5% (13).
Staphylococcus epidermidis is among the most prevalent pathogens implicated in nosocomial bloodstream infections. The incidence of infections associated with permanent implants, such as vascular grafts, prosthetic joints, and cardiac devices, ranges from 1% to 3%. Staphylococcus epidermidis accounts for approximately 30 - 43% of prosthetic joint infections, around 22% of bloodstream infections in intensive care units, and about 13% of prosthetic valve infections. It is also a leading cause of neonatal sepsis. These infections contribute to prolonged hospital stays, the need for additional surgical interventions, and increased mortality rates (14, 15).
Managing S. epidermidis infections presents significant challenges due to the species' high levels of antibiotic resistance and its ability to form biofilms (16). Probiotics may impact S. epidermidis through various mechanisms, including competitive exclusion, production of inhibitory substances such as organic acids and bacteriocins, modulation of the host immune response, and disruption of biofilm formation (17).
2. Objectives
This study aimed to investigate the effectiveness of isolated probiotics in inhibiting the growth of S. epidermidis.
3. Methods
3.1. Isolation of Staphylococcus epidermidis
In this cross-sectional study, S. epidermidis was utilized as an in-vitro model (18). The bacteria were cultured and identified through microbiological and biochemical tests (19).
3.2. Antibiotic Susceptibility Test
Following the CLSI guidelines, the disc diffusion method was employed to determine the antibiotic resistance patterns of clinical isolates. The antibiotics used in the study included levofloxacin (5 μg), ciprofloxacin (5 μg), meropenem (10 μg), imipenem (10 μg), piperacillin-tazobactam (100-10 μg), amikacin (30 μg), gentamicin (10 μg), ampicillin-sulbactam (10 μg), ceftazidime (30 μg), and cefepime (30 μg) (BD, USA).
3.3. Isolation of Probiotics Strains from Local Dairy Samples
A total of 10 samples, comprising locally sourced milk (cow milk), yogurt (cow milk), and curd, were utilized to isolate probiotics. These samples were collected from various regions within Isfahan, Iran. To isolate the bacteria, 1 mL of each dairy sample was mixed with a 2% w/v sodium citrate solution and homogenized. Subsequently, 1 mL of the prepared samples was added to 10 mL of MRS broth and incubated at 37°C for 24 hours. Following the incubation period, 0.02 mL of the solutions were spread onto MRS agar media and allowed to grow for 48 hours. The identification of strains involved conducting a catalase test, Gram staining, and biochemical tests, including evaluating growth at temperatures of 15 and 45°C, assessing acid and gas production from glucose, observing NH3 production from arginine, and examining sugar fermentation capabilities for arabinose, cellobiose, mannitol, mannose, melebiose, raffinose, ribose, salicin, rhamnose, and xylose (20).
3.4. Assessment of Antibacterial Activity of Probiotics Strains
We conducted agar well diffusion and broth microdilution tests to assess the antibacterial activity of the probiotics (21).
3.5. Agar Well Diffusion Method
We extracted the cell-free supernatant from the probiotic cultures and utilized it in the agar well diffusion method (20). Antibacterial activity was assessed by measuring the zones of growth inhibition surrounding the wells after 24 hours of incubation at 37°C.
3.6. Broth Microdilution Assay
A broth microdilution test was conducted to assess the antibacterial activity, minimum inhibitory concentration (MIC), and minimum bactericidal concentration (MBC) of the cell-free supernatant from probiotics against clinical isolates of S. epidermidis. A series of dilutions (1/2, 1/4, 1/8, and 1/16) of the Lactiplantibacillus cell-free supernatant was prepared and added to 96-well plates containing MRS broth medium. The plates were inoculated with a suspension of the clinical isolates of S. epidermidis (final inoculum approximately 106 CFU/mL) and incubated at 37°C for 24 hours. Subsequently, the contents were cultured on blood agar medium and incubated again at 37°C for 24 hours (20, 22). Negative controls were included in the experiment. The MIC and MBC values were determined by measuring the optical density at 470 nm (OD470 nm) (23).
3.7. Time-Kill Test in Co-cultures
The time-kill test was conducted as previously described, with some modifications (20). Probiotic colonies were inoculated into tubes containing MRS broth medium and incubated for 72 hours. Subsequently, 2 mL of the supernatant was transferred into autoclaved sterile tubes. Using sterile loops, the pathogenic colony was inoculated into physiological serum to prepare a suspension with 0.5 McFarland turbidity (1.5 × 108 CFU/mL). Then, 400 µL of this suspension was added to tubes containing 2 mL of the supernatant. Bacterial growth or absence was observed on blood agar medium at 0, 24, 48, 72, and 120 hours to evaluate bacterial growth.
3.8. Assessment of Acid Tolerance of Probiotic
The probiotic strains were cultured in MRS broth for 48 hours at 37°C. Following this, they were transferred into PBS solutions adjusted to varying pH levels (pH 1, pH 2, pH 3, and pH 4 as a control) and incubated at 37°C for 0 minutes, 30 minutes, and 1 hour. After the incubation periods, the bacterial suspensions were plated onto MRS agar and incubated for 24 hours at 37°C. The survival rates of the probiotics under normal and acidic conditions (pH 1, pH 2, and pH 3) were evaluated to assess their acid tolerance. This experimental procedure was performed in duplicate to ensure accuracy.
3.9. Anti-biofilm Effect of Probiotics Using Microtiter Plate Test
Bacterial isolates were cultured in MRS medium for 24 hours at 37°C. Suspensions with a density equivalent to a 0.5 McFarland standard were prepared and inoculated into MRS medium supplemented with 0.2% sucrose, along with cell-free supernatant extracts at concentrations of 1/2, 1/4, 1/8, and 1/16 of the MIC values. Subsequently, 200 μL of the prepared solutions was dispensed into each well of a 96-well microplate. Wells without microorganisms and cell-free supernatant served as positive and negative controls, respectively. The microplate was incubated at 37°C for 24 hours. Following incubation, crystal violet staining was performed, the wells were washed with 95% ethanol, and the optical density (OD) was measured at 570 nm. This procedure was repeated three times, using positive controls (S. epidermidis) and negative controls (uninfected medium), to ensure the reliability of the results.
3.10. Determining the Possible Inhibitory Mechanism
Probiotics initially adjusted to pH 4 were neutralized to pH 7 by adding four drops of sodium hydroxide (NaOH). The probiotics were then divided into two groups: One containing neutralized probiotics (pH 7) and the other containing the original probiotic supernatant (pH 4). The agar well diffusion method on Muller Hinton agar was utilized as described previously. Plates were inoculated with S. epidermidis culture standardized to a turbidity of 0.5 McFarland. Using a sterile glass Pasteur pipette, four wells were created on each plate. Two wells were filled with neutralized probiotics (pH 7), while the other two were filled with the original probiotics (pH 4). The plates were incubated at 37°C for 24 hours.
Simultaneously, a suspension of S. epidermidis with a turbidity of 0.5 McFarland was prepared in a volume of 1000 μL. An equal volume of neutralized probiotics was added to this suspension in a sterile screw-capped tube. After 4 hours of incubation, 50 μL of the mixture were inoculated onto chocolate agar medium. The inoculated plates were further incubated at 37°C for 24 hours.
3.11. High-Performance Liquid Chromatography
The probiotic strains were cultivated for 72 hours in MRS broth medium. The supernatant was carefully separated and filtered through a 0.25 μm syringe filter to ensure purity. High-performance liquid chromatography (HPLC) analysis was performed on the cell-free supernatant. The flow rate was maintained at 1 mL/min, and the pH was set to 3.6 during the analysis. The mobile phase consisted of an aqueous solution containing 10 mM phosphate buffer and acetonitrile. The analysis utilized reversed-phase HPLC columns (C18, 25 cm × 4.6 mm) for the separation and detection of compounds (24).
3.12. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide Assay
The cytotoxic effects of the CFS on normal subcutaneous connective tissue cells (L929) were evaluated using a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, as described in previous studies (25). The L929 cell line was cultured in low-glucose DMEM supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (streptomycin and penicillin) at 37°C in a humidified atmosphere containing 5% CO2, using a 96-well microplate for cell culture.
Each well was filled with 100 µL of culture medium. The cells were treated with a filtered probiotic supernatant diluted in dimethyl sulfoxide (DMSO) and culture medium in the first well of each row. A serial dilution of the compounds was prepared for the remaining wells, with concentrations ranging from 3.12% to 50%.
Afterward, 100 µL of a cell suspension at a density of 1 × 104 cells per well was added to each well. A control well containing only cells with culture medium and a blank well containing solely culture medium were included. The microplate was incubated for 24 hours. Post-incubation, the wells were washed three times with culture medium supplemented with FBS to remove any residual compounds or substances. Subsequently, 20 µL of MTT reagent (5 mg/mL in sterile PBS) was added to each well, and the plate was incubated at 37°C for three hours. The cells' ability to metabolize MTT to formazan crystals was then evaluated.
The resulting crystals were dissolved with 100 µL of DMSO solution per well, and the plate was gently agitated for 30 minutes to ensure complete dissolution. Following the removal of the culture media, the absorbance was measured using a Bio-Rad microplate reader (California, USA) at a wavelength of 570 nm (7).
The assay was conducted in triplicate, and results were expressed as the mean ± standard deviation (SD). Cell viability was calculated as a percentage relative to the control group using the following formula:
%Cell viability = [(Sample absorbance - Blank absorbance)/(Cell growth control absorbance - Blank absorbance)] × 100
3.13. Identification of Selected Probiotics
DNA extraction was carried out from pure bacterial cultures (7). Bacterial identification was confirmed using traditional biochemical tests, and universal primers were obtained from CinnaGen Co., Iran. Amplification of the 16S rRNA gene was performed using the following PCR protocol: An initial denaturation step at 95°C for 5 minutes, followed by 30 cycles of 30 seconds denaturation at 95°C, 30 seconds annealing at 54°C, and 30 seconds extension at 72°C, with a final extension step at 72°C for 5 minutes.
Direct sequencing of the PCR products was outsourced to SinaClon, Iran, for nucleotide sequence determination. The resulting sequences were compared with NCBI's BLAST database for identification and were subsequently documented.
3.14. Statistical Analysis
The statistical analysis was conducted using SPSS™ software, version 16 (IBM Corp., USA). Categorical variables were expressed as counts and percentages. Significant differences were assessed using either Fisher's exact test or the chi-square (χ2) test. A P-value of less than 0.05 was considered statistically significant.
4. Results
4.1. Staphylococcus epidermidis Isolation
In this study, a specimen of S. epidermidis isolated from a urine sample was utilized.
4.2. Probiotic Isolation and Identification
Two probiotic strains were isolated from dairy samples, including yogurt and milk, to examine their antibiofilm and antibacterial properties against S. epidermidis strains. The cell-free supernatants of these two probiotic strains, designated as 3A and 5A, exhibited inhibitory effects, as evidenced by inhibition zones on S. epidermidis strains. Both the 3A and 5A strains were derived from dairy samples. Moreover, acid tolerance tests indicated that strains 3A and 5A demonstrated resistance to acidic conditions. Biochemical analyses were conducted to identify these two probiotics, 3A and 5A.
4.3. Identification of Selected Probiotic, Sequencing, and Registration in NCBI
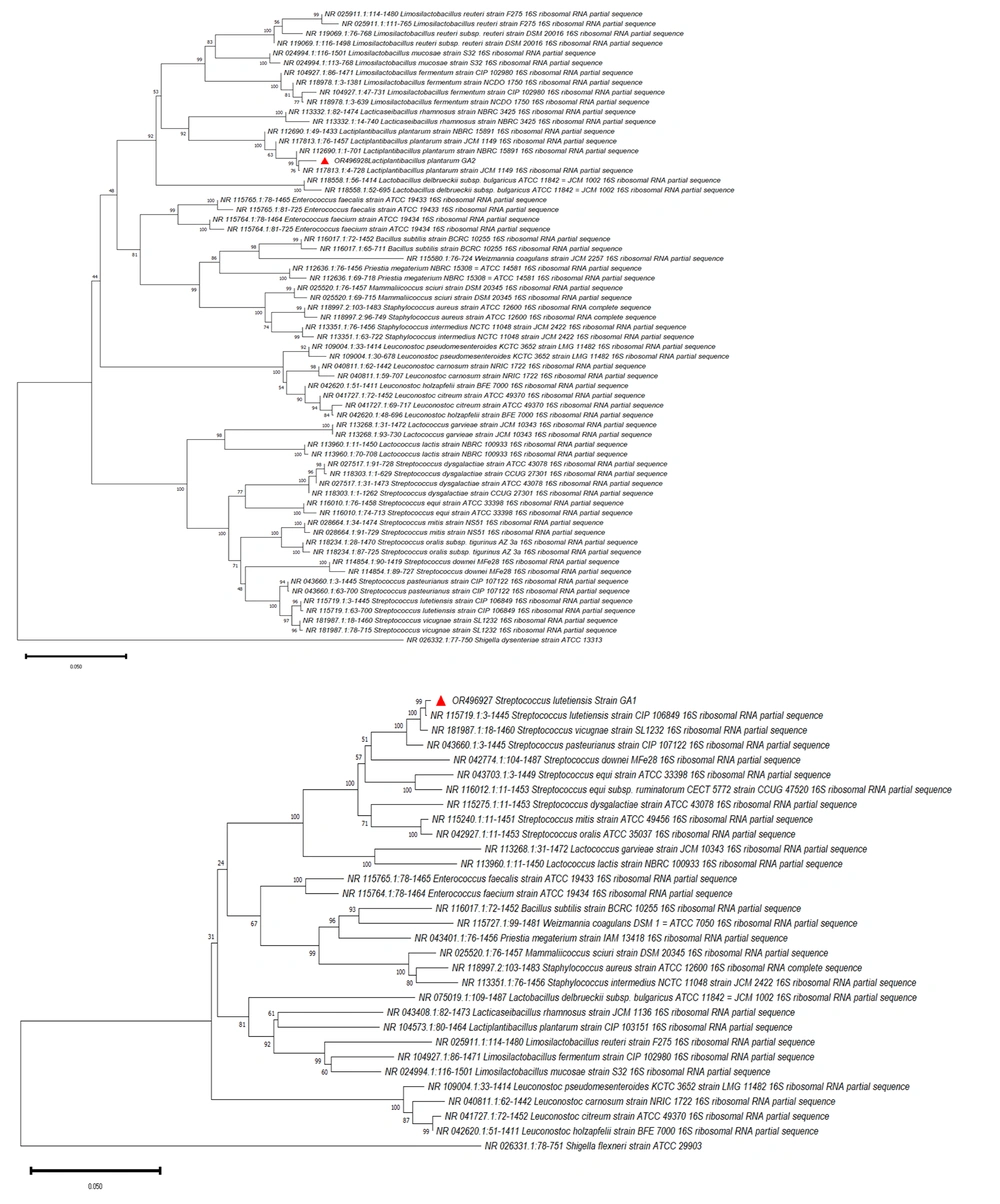
Two probiotic strains, 3A and 5A, which exhibited the highest inhibitory activity, were identified. The PCR products were sequenced and analyzed, with gel electrophoresis performed for confirmation. Biochemical tests and BLAST sequence analysis of the 16S rDNA revealed that strain 3A belonged to Streptococcus lutetiensis OR496927.1, and strain 5A belonged to Lactiplantibacillus plantarum OR496928, as corroborated by the available data in the sequence database. The genome sequences of these bacteria were submitted to the NCBI database, and a phylogenetic tree was constructed (Figure 1).
4.4. Antimicrobial Effect of Probiotics against Staphylococcus epidermidis
Among the two probiotic strains, the cell-free supernatant of a particular strain exhibited notable inhibitory effects, as demonstrated by inhibition zones measuring 14 mm in diameter against all tested strains of S. epidermidis in the agar well diffusion method, based on the obtained results. The MIC for L. plantarum was 1/4, and the MBC was 1/2. For S. lutetiensis, the MIC was 1/2, and the MBC was 1.
4.5. Assessment of Acid Tolerance of Probiotic
The L. plantarum strain demonstrated growth at pH 1 and pH 2 at zero time, but no growth was observed at these pH levels after 30 minutes and one hour. However, the L. plantarum strain exhibited growth at pH 3 at all-time points: Zero time, 30 minutes, and one hour. In contrast, the S. lutetiensis strain showed no growth at pH 1 and pH 2 across all time points but demonstrated growth at pH 3 and pH 4.
4.6. Determining the Possible Inhibitory Mechanism
The investigation into the potential inhibition mechanism of L. plantarum and S. lutetiensis strains revealed that their cell-free supernatants produced inhibition zones against S. epidermidis isolates in the agar well diffusion method at pH 4. However, neutralized supernatants (adjusted to pH 7) from both strains showed no inhibitory activity against S. epidermidis, indicating that the inhibitory effects of L. plantarum and S. lutetiensis strains were likely due to the production of organic acids.
4.7. Anti-biofilm Activity of Probiotic
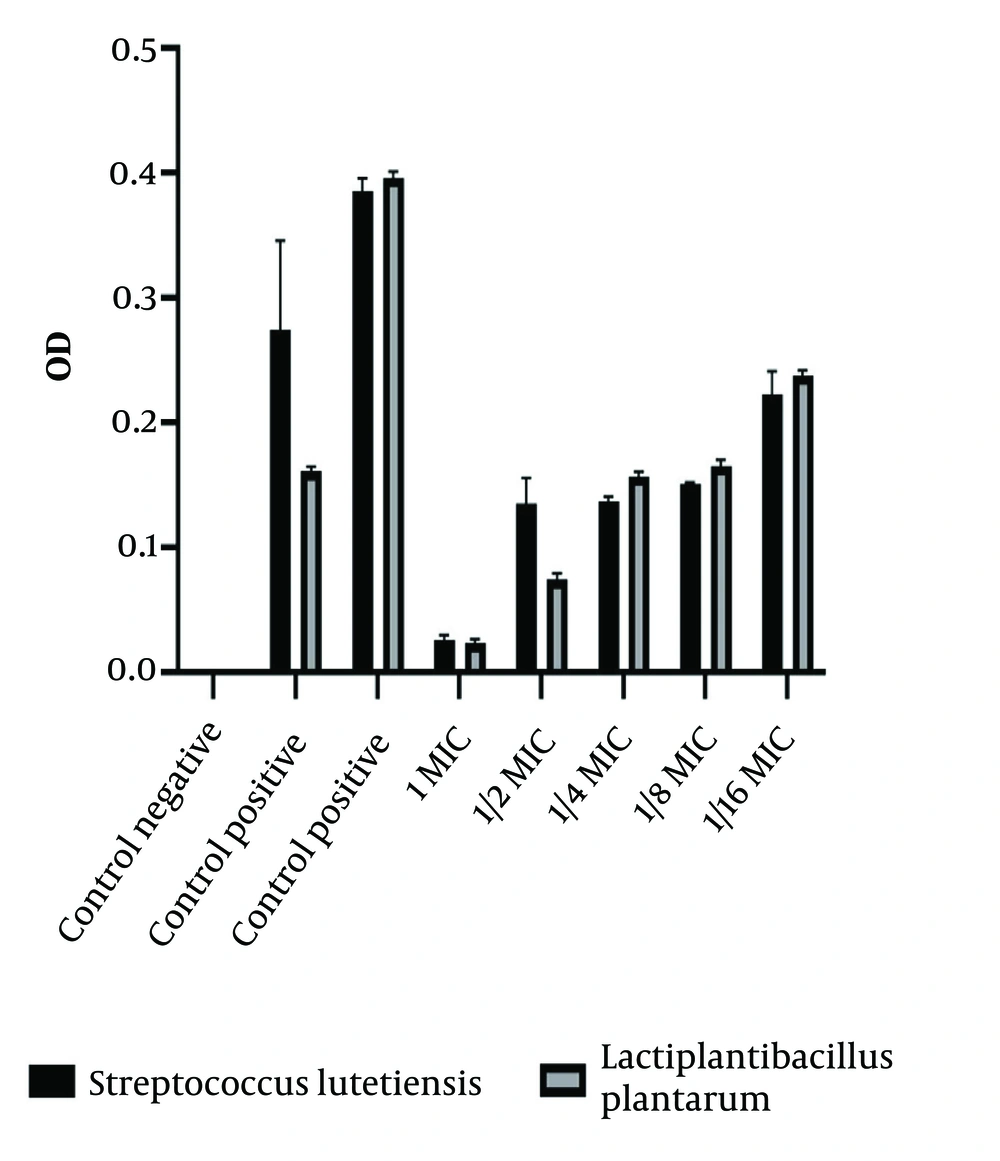
The results showed that after the addition of crystal violet and 98% ethanol, no coloration was observed in the wells where the probiotic exhibited anti-biofilm activity, indicating the prevention of biofilm formation by the pathogenic strain. In contrast, visible coloration in other wells suggested that the pathogenic strain had formed a biofilm and adhered to the surfaces, and the probiotic strain could not inhibit this formation. A positive control of the probiotic strain confirmed its ability to form a biofilm as well (Figure 2).
The results of the anti-biofilm activity of S. lutetiensis and L. plantarum strains against the positive control pathogen showed varying levels of inhibition at different concentrations. S. lutetiensis demonstrated moderate inhibitory effects at concentration 1 (0.224) and more potent inhibition at concentration 1/2 (0.393), with reduced effectiveness at lower concentrations. The L. plantarum strain exhibited mild inhibition at concentration 1 (0.159) and stronger inhibition at concentration 1/2 (0.780), followed by reduced effectiveness at concentrations of 1/4 and lower.
Overall, both strains displayed anti-biofilm activity, indicating their potential as probiotics to combat biofilm formation. Among the two, the L. plantarum strain generally showed stronger inhibitory effects than the S. lutetiensis strain, highlighting its greater potential for use in biofilm-related applications.
4.8. HPLC Results
The HPLC results revealed that lactic acid, formic acid, and acetic acid were the predominant organic acids produced by all probiotic strains. The concentrations of these acids were as follows: Lactic acid at 2.60 g/100 g, formic acid at 0.04 g/100 g, and acetic acid at 0.60 g/100 g.
4.9. Cell Viability Results
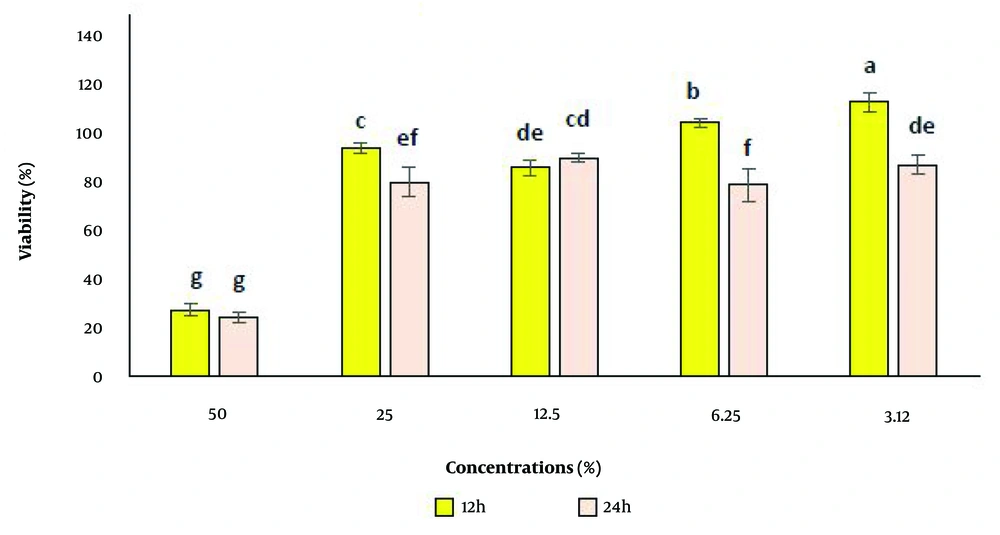
The percentage of cell viability in contact with the test sample, compared to the control sample, was assessed at five different concentrations (3.12%, 6.25%, 12.5%, 25%, and 50%). Cell viability was evaluated after 24 ± 2 hours and 12 ± 2 hours of exposure to the original sample and control, with three repetitions conducted for accuracy. Toxicity was observed at a concentration of 50% after both time intervals, while cell viability remained unaffected at the lower concentrations (Figures 3 and 4).
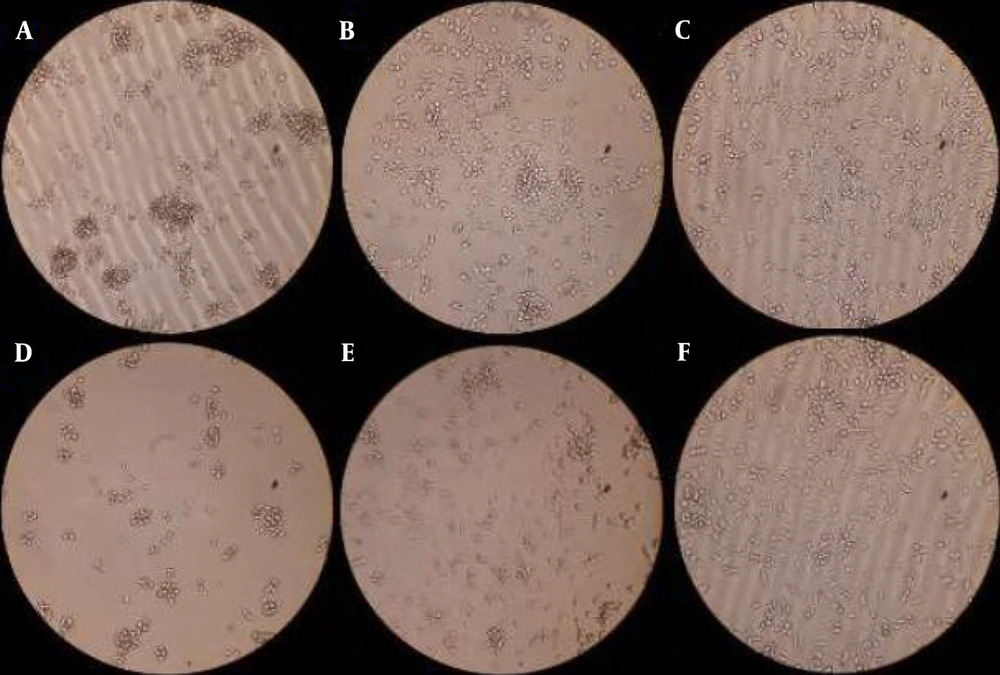
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) results of applying Streptococcus lutetiensis and Lactiplantibacillus plantarum on Staphylococcus epidermidis. At two concentrations 50 and 3.12% of 3B sample and control sample after 24 ± 2 and 12 ± 2 hours: A and B, concentration of 3.12% and 50% of 3B sample after 12 ± 2 hours; C, control sample after 12 ± 2 hours; D and E, concentration of 3.12% and 50% of 3B sample after 24 ± 2 hours; F, control sample after 24 ± 2 hours.
5. Discussion
The increasing prevalence of antibiotic resistance and the recognition of the human microbiota's significant role in health and disease have amplified interest in probiotic therapies as alternative or adjunctive strategies to address bacterial infections (26-28). While probiotics are widely regarded as beneficial, the observed cytotoxicity highlights the need for careful evaluation when determining the optimal dosage and duration of probiotic use. This study focused on isolating and identifying probiotics effective against S. epidermidis-induced UTIs. Although S. epidermidis is a common skin commensal, its presence in urine samples from hospitalized patients, particularly the elderly, poses a diagnostic challenge. Distinguishing between benign colonization and true infection is essential, especially in non-catheterized patients. Staphylococcus epidermidis is a frequent cause of UTIs, particularly those associated with catheters (29). John et al. reported a 10.2% prevalence of S. epidermidis in urine samples (30). Most S. epidermidis infections occur in conjunction with indwelling medical devices such as urinary catheters or in immunocompromised patients. Community-acquired S. epidermidis UTIs are rare, especially in children, and healthcare providers often overlook this pathogen as a causative agent due to its antibiotic resistance and biofilm formation capabilities (31).
In this study, the probiotics Streptococcus lutetiensis and Lactiplantibacillus plantarum were isolated from dairy samples. Some strains within the Streptococcus bovis/Streptococcus equinus complex, such as S. lutetiensis and S. gallolyticus, are frequently derived from dairy products and are considered safe probiotics (32, 33). Jaleel and Kiliç isolated L. plantarum and L. fermentum from dairy and fermented foods, demonstrating that L. plantarum had the highest adhesion percentage and was non-toxic at a concentration of 108 CFU/mL (34). Jaleel and Kiliç confirmed the antimicrobial activity of L. plantarum, attributing its effects to the production of organic acids and hydrogen peroxide (34).
This study observed that S. lutetiensis and L. plantarum strains exhibited cytotoxic effects on S. epidermidis isolates at 50% and 25% concentrations, respectively. These findings suggest that L. plantarum is effective at lower concentrations for inhibiting S. epidermidis growth, while S. lutetiensis is more effective at lower concentrations for bacterial elimination. Selegard et al. also reported that L. plantarum demonstrated efficacy against S. epidermidis when used in combination with low doses of conventional antibiotics (35).
Lactiplantibacillus plantarum is distinguished for its versatility and extensive application as a lactic acid bacterium. Its efficacy as a potent probiotic biopreservative surpasses that of many other Lactobacillus species, largely due to its dual role as a natural resident of the human gut and a well-documented starter culture in food fermentation processes, ensuring its prolonged and safe usage (36). In our study, both L. plantarum and S. lutetiensis exhibited anti-biofilm activity, with L. plantarum demonstrating stronger effects. Carvalho et al. similarly reported that L. plantarum caused significant biofilm inhibition, achieving a 76% reduction after 24 hours (37). Lactobacillus species, whether in lyophilized or fermented forms, act as probiotics that shield the host against harmful microorganisms, enhance immune system functionality, improve feed digestibility, and mitigate metabolic disorders (38, 39). Our findings revealed that L. plantarum could thrive at pH 3, while S. lutetiensis exhibited growth at both pH 3 and 4. Ingham et al. corroborated that the optimal pH for L. plantarum growth is approximately pH 3, demonstrating the resilience of a subpopulation of cells at this pH after a rapid pH downshift from 2 to 4, although growth in liquid culture was inhibited (40). Similarly, studies have shown that the growth of Enterococcus avium, S. equinus, and S. lutetiensis, isolated from the rumen, was inhibited at pH 4 (41).
In this study, lactic acid, formic acid, and acetic acid were identified as the predominant organic acids produced by S. lutetiensis and L. plantarum. The inhibitory effects of these strains were attributed to their production of organic acids, as neutralized supernatants (pH 7) did not exhibit inhibitory activity. The production of organic acids varies across bacterial strains and is influenced by culture composition and growth conditions (42).
Previous research has consistently confirmed the inhibitory effects of organic acids produced by Lactobacillus strains against pathogenic bacteria (20). For instance, Ghiaei et al. demonstrated that lactic acid produced by L. rhamnosus exhibited inhibitory effects against Pseudomonas aeruginosa (7). Similarly, Abbasi et al. found that lactic acid and acetic acid produced by L. rhamnosus inhibited the growth of Acinetobacter baumannii (10). Furthermore, Shokri et al. reported that organic acids, including lactic acid, acetic acid, and formic acid, produced by a bacteriocin-negative strain of Lactobacillus fermentum, suppressed the growth of P. aeruginosa (20). According to Mohamed et al., cell-free preparations of probiotics, particularly Lactobacillus acidophilus EMCC 1324, exhibited antibacterial effects against certain antibiotic-resistant strains of Staphylococcus aureus and S. epidermidis. These findings suggest potential applications, provided adequate safety evidence is established (43). Kheirjou et al. examined the impact of Bacillus coagulans supernatant, a spore-forming probiotic, on the formation of persister cells of S. epidermidis. Their study revealed that the supernatant, containing bacterial metabolites, significantly reduced the number of persister cells at high concentrations (44).
The inhibitory effects of postbiotics are largely attributed to the production of bacteriocins and organic acids. A strong correlation has been identified between the levels of organic acids, such as acetic acid, lactic acid, and caproic acid, and inhibitory activity against pathogens. This suggests that organic acid production enhances the activity of bacteriocins (45). Lactic acid and other organic acids lower pH levels, thereby inhibiting microbial growth. However, probiotics remain unaffected due to their tolerance to low pH environments (46). Our HPLC analysis indicated that the inhibitory activity of S. lutetiensis and L. plantarum strains against S. epidermidis was primarily mediated by the production of these organic acids. This study had several limitations. Firstly, in vivo studies are essential to confirm the therapeutic potential and safety of these probiotics within the human body. Secondly, while this research demonstrated the antibacterial and anti-biofilm properties of probiotics against antibiotic-resistant S. epidermidis, clinical trials are needed to evaluate these effects in real-world patient scenarios. Clinical studies could provide valuable insights into appropriate dosages, potential side effects, and effectiveness across diverse patient populations—factors beyond the scope of this in vitro study.
5.1. Conclusions
Our study demonstrated the efficacy of the probiotics S. lutetiensis and L. plantarum, isolated from dairy samples, against S. epidermidis infections. Both strains exhibited strong acid tolerance and significant anti-biofilm activity, primarily attributed to their production of organic acids. Importantly, these probiotics showed a favorable safety profile, as evidenced by the cell viability results. These findings underscore the therapeutic potential of S. lutetiensis and L. plantarum as promising probiotics for combating S. epidermidis infections.