1. Background
Capparis spinosa, also known as the caper bush, is a medicinal plant that has been used in traditional medicine for thousands of years. The plant is native to the Mediterranean region and parts of Central Asia and has been employed to treat a wide range of conditions, including rheumatism, headaches, toothaches, and skin diseases (1, 2). In Iran, C. spinosa grows widely across several provinces and is known by the common name "Alaf-e-Mar" (3). Its fruits, leaves, roots, and bark have been utilized in Iranian traditional medicine to alleviate fever, gastrointestinal disorders, liver diseases, hemorrhoids, and gout (4).
The phytochemistry of C. spinosa has been extensively studied, and various bioactive components have been identified in the aerial parts, fruits, seeds, and roots of the plant. These include compounds such as flavonoids, alkaloids, glucosinolates, fatty acids, and sterols. The fruits contain high levels of antioxidant flavonoids like rutin, quercetin, and kaempferol glycosides. Alkaloids isolated from the fruits include capparisine A, B, and C, along with derivatives like stachydrine. The seeds are a rich source of glucosinolates, predominantly glucocapperin, as well as sterols like β-sitosterol and δ-tocopherol. They also contain high percentages of beneficial unsaturated fatty acids like oleic and linoleic acid (5).
Recent scientific research has explored several therapeutic effects of C. spinosa extracts and isolated constituents. Studies have demonstrated the antioxidant and antidiabetic activities of caper fruit extracts in animal models as well as diabetic patients. Aqueous and methanolic extracts have also exhibited anti-inflammatory effects in vivo by reducing edema, immune cell infiltration, and expression of inflammatory cytokines (6-8). The antimicrobial effects of C. spinosa against bacteria like Staphylococcus aureus and fungi like Aspergillus flavus have been shown using in vitro bioassays (9). While there is evidence for diverse therapeutic effects, the specific modes of action and molecular targets underlying the bioactivity of C. spinosa still need to be fully elucidated. Network pharmacology approaches that model compound-target interactions and biochemical pathways promise to provide greater insights into the multi-target mechanisms of medicinal herbs like the caper bush.
2. Objectives
The present study aims to predict the targets and pharmacological actions of traditionally used C. spinosa by using integrated network pharmacology strategies like compound-target network analysis. Molecular docking will also be performed to verify the reliability of the network predictions. This network-based investigation will shed light on the therapeutic potential and diverse bioactivities exhibited by pharmacological research on C. spinosa.
3. Methods
3.1. Collection and Screening of Active Compounds of Capparis spinose
The known phytochemical compounds present in C. spinosa were collected from a literature review of published research articles on C. spinosa. Key articles searched included the Annaz et al. (2) and Zhang and Ma (10) review articles. The qualitative and quantitative descriptors of oral bioavailability (OB) and drug-likeness (DL) were obtained from Swiss ADME (11) and ADMET LAB 2.0 (12). We used OB and DL thresholds to ensure the compounds have potential for therapeutic use. The OB threshold of > 30% was chosen because it is a widely accepted cutoff in pharmacokinetic studies to identify compounds with good absorption and bioavailability. Similarly, the DL threshold was based on Lipinski’s rule of five, which is a standard in drug discovery to predict compounds with favorable pharmacokinetic properties. The 2D structures, molecular formula, and the canonical SMILES of the 52 screened compounds were downloaded from the PubChem database.
3.2. Prediction and Screening of Targets of Capparis spinosa Active Components
The protein targets of the active compounds from Capparis spinosa were predicted using three complementary approaches:
1. SwissTargetPrediction: The canonical SMILES format of each compound was submitted to the SwissTargetPrediction web server (http://www.swisstargetprediction.ch/), with Homo sapiens selected as the target organism. Results were filtered to retain only targets with a probability score greater than 0.7.
2. PharmMapper Server: Compound structures in MOL2 format were uploaded to the PharmMapper tool (http://59.78.96.61/pharmmapper), using the "Human Protein Targets Only" option. Targets were filtered based on a normfit score ≥ 0.8 and a fit score ≥ 3, with the top 10 targets retained for each compound.
3. BindingDB Database: BindingDB uses a similarity-based approach to predict protein targets of small-molecule ligands. A similarity threshold of 85% was applied, and all available human protein targets for each query compound were retrieved. The targets identified from all three platforms were consolidated and mapped to their official gene symbols using UniProt ID mapping. Redundant entries were removed to generate a non-redundant list of protein targets. The relationships between the compounds and their predicted targets were visualized as a network using Cytoscape version 3.10.1 (https://cytoscape.org)
3.3. The Gene Ontology and Enrichment Analysis of Targets of Capparis spinosa
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis of the 183 C. spinosa targets was conducted using the bioinformatics.com.cn.en database. The enriched GO terms were categorized into three main categories: Biological process, molecular function, and cellular component. For pathway analysis, the gene list was searched against the KEGG pathway database. Significantly enriched pathways were identified based on an adjusted P-value threshold of < 0.05. Further prioritization was done based on the combined score, which indicates the degree of enrichment.
3.4. Identification of the Cancer-Related Target Genes
A list of cancer-related genes was obtained by querying the KEGG database using the search term "cancer". The combined list resulted in 1852 cancer-associated genes, which were used for further analysis after duplicate removal.
3.5. Protein-Protein Interaction Network of the Cancer Targets to Introduce Hub Targets
Protein interaction analysis of the non-redundant cancer targets was conducted using the STRING database version 11.5. A confidence threshold of 0.7 (high confidence) was set for protein-protein interaction (PPI) retrieval, and the active interaction sources were based on text mining, experiments, and databases. The data from the resultant interaction network were imported into Cytoscape and analyzed to define network topological properties such as degree centrality, betweenness centrality, and closeness centrality. Based on degree centrality, the cancer targets were ranked, and nodes with the highest degree were designated as hub targets.
3.6. Common Target Identification of Cancer Related Targets with Capparis spinosa Targets
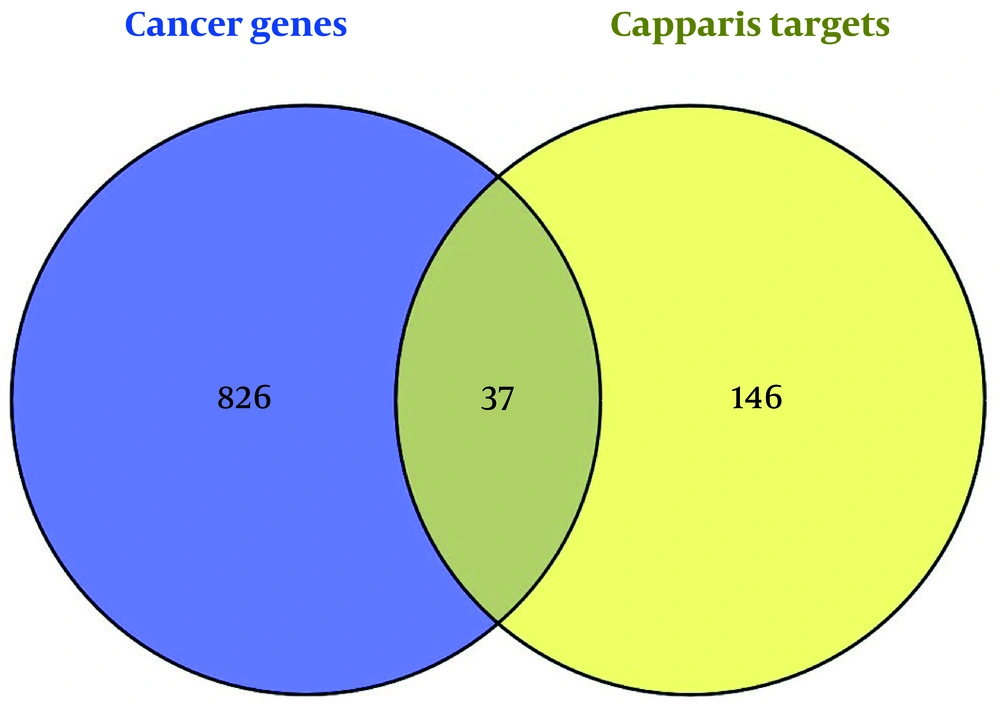
The cancer-associated gene list and the C. spinosa target list were compared using the Venny 2.1 tool to identify common targets. This comparison revealed an overlap of 37 genes that may play a role in the anticancer activity of C. spinosa.
3.7. Mapping Common Targets on the Kyoto Encyclopedia of Genes and Genomes Cancer Pathways
The 37 common targets between cancer and C. spinosa were mapped onto specific KEGG cancer-associated pathways, such as Pathways in Cancer, p53 Signaling, and Cell Cycle, using the KEGG Mapper visualization tool. This mapping provides a representation of where these targets are involved in the pathway diagrams, offering insights into the potential mechanisms through which C. spinosa may exert its anticancer effects.
3.8. Molecular Docking to Validate Binding Affinity
Molecular docking was performed to evaluate the binding interactions between the active compounds of C. spinosa and the identified hub targets (AKT1, EGFR, and SRC). The docking simulations were carried out using AutoDock Tools (version 1.5.6), a widely used software for predicting ligand-protein interactions (13). The 3D structures of the target proteins (AKT1: PDB ID 3O96, EGFR: PDB ID 1M17, and SRC: PDB ID 2SRC) were obtained from the Protein Data Bank (PDB). Water molecules and co-crystallized ligands were removed, and hydrogen atoms were added to the proteins using AutoDock Tools. The protein structures were then prepared by assigning Gasteiger charges and merging non-polar hydrogens.
The 2D structures of the C. spinosa compounds were converted into 3D formats using Open Babel, and energy minimization was performed using the universal force field (UFF) in Avogadro software. The ligands were prepared by adding Gasteiger charges and setting rotatable bonds. A grid box of 60 × 60 × 60 Å with a spacing of 0.375 Å was created around the active site of each protein to ensure comprehensive coverage of potential binding pockets. The Lamarckian genetic algorithm (LGA) was used for docking simulations, with 100 runs per ligand to explore different binding conformations. The docking parameters included a population size of 150, an energy evaluation of 2,500,000, and a maximum of 27,000 generations.
The best binding pose for each ligand was selected based on the lowest binding energy (ΔG, kcal/mol) and inhibition constant (Ki). The binding interactions, including hydrogen bonds, hydrophobic contacts, and van der Waals forces, were visualized and analyzed using LigPlot+ (version 2.2). Validation of the docking protocol was performed by re-docking co-crystallized ligands and comparing them with reference inhibitors.
4. Results
4.1. Active Ingredients and Related Targets of Capparis spinose
Among the screened compounds, 31 compounds had predicted targets based on our cutoffs. The canonical SMILES, OB, and DL values of these compounds are presented as Appendix 1 in Supplementary File. After removing redundant targets, 183 non-redundant protein targets were identified and used for further analyses. We conducted a compound-target network to illustrate the targets associated with each compound (Figure 1). The network included 31 compound nodes, 183 target nodes, and a total of 863 degrees between them. The degree of each compound node was mapped to its size in the network. As shown in Figure 1, apigenin, kaempferol, and gallic acid had the highest degrees with 65, 60, and 32 targets, respectively.
4.2. The Gene Ontology and Enrichment Analysis Results
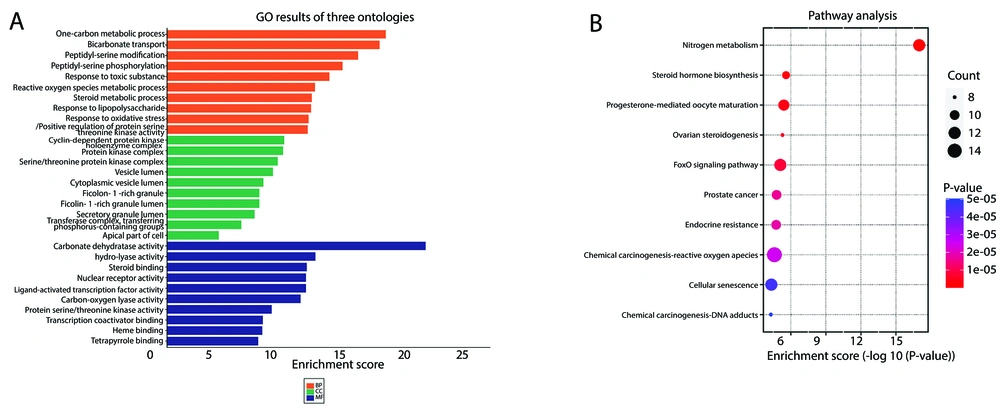
The ontologies of the identified targets of C. spinosa, including biological processes, cellular compartments, and molecular functions, are shown in Figure 2A. It can be seen that the targets were associated with main regulatory processes such as protein modification, phosphorylation, steroid metabolism, and regulation of inflammatory/oxidative response. The predicted targets were enriched in specific KEGG pathways. As depicted in Figure 2B, our targets were markedly involved in nitrogen metabolism, chemical carcinogenesis, cancer, FoxO signaling pathway, and steroid hormone biosynthesis pathways among the top 10 records.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis. A, GO functional enrichment analysis (showing the top 10 entries only). B, KEGG pathway enrichment analysis (the top 10 entries shown only).Abbreviations: BP, biological processes; CC, cellular components; MF, molecular functions.
4.3. The Cancer-Related Target Genes and Their Protein-Protein Interaction
A total of 1852 cancer-related genes were recovered from KEGG, which were shortlisted to 863 non-redundant genes. The STRING database provided the cancer genes interaction network containing 695 nodes and 8541 edges. The network was further analyzed using Cytoscape tools, and the topological properties were recovered.
4.4. Intersecting Targets and Their “Hub-Targets”
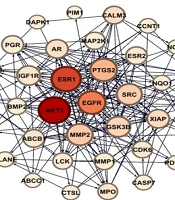
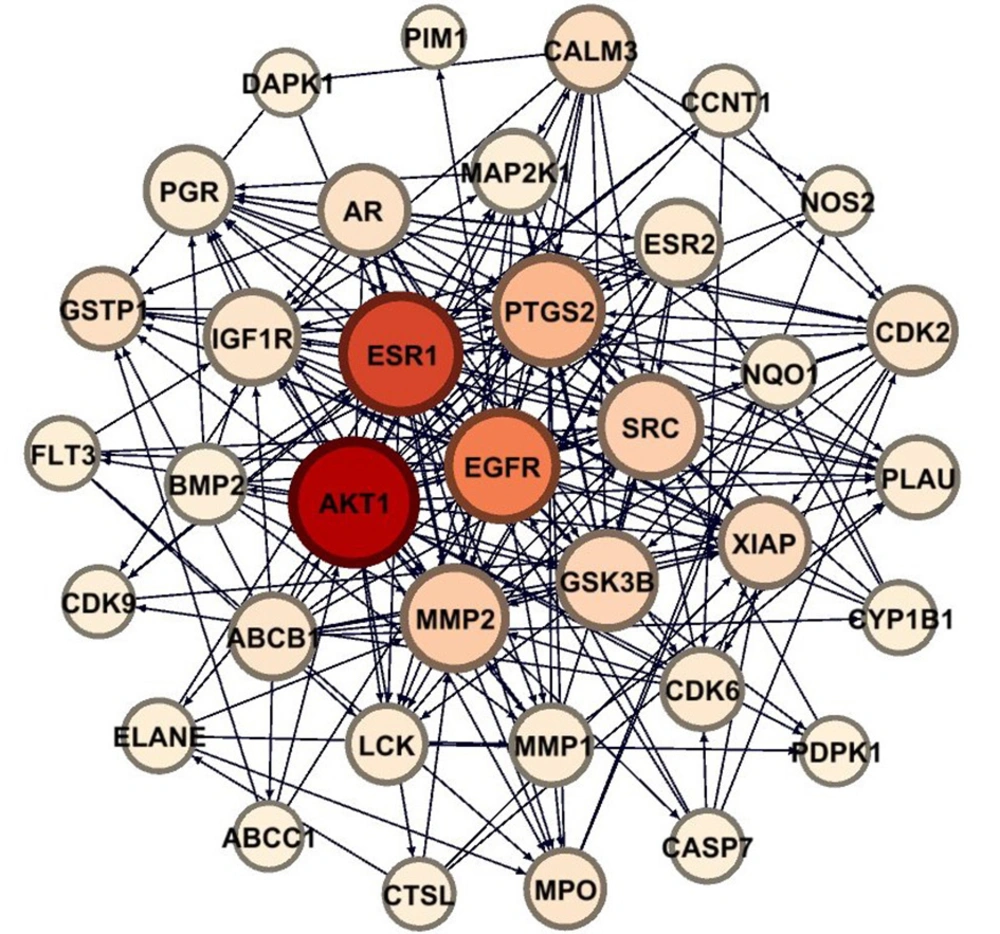
Analysis using the Venny 2.0 tool identified 37 intersecting targets between the predicted compound targets and known cancer-related genes (Figure 3), with their interactions visualized in Figure 4. These intersecting targets were mapped onto the cancer gene network to evaluate their positions and network parameters, as detailed in Appendix 2 in Supplementary File.
Hub genes were defined as nodes with the highest connectivity, specifically those with a node degree greater than 100. According to the supplementary table of intersecting targets, three predicted targets — AKT1, EGFR, and SRC — were identified as hub genes, with node degrees of 185, 170, and 163, respectively.
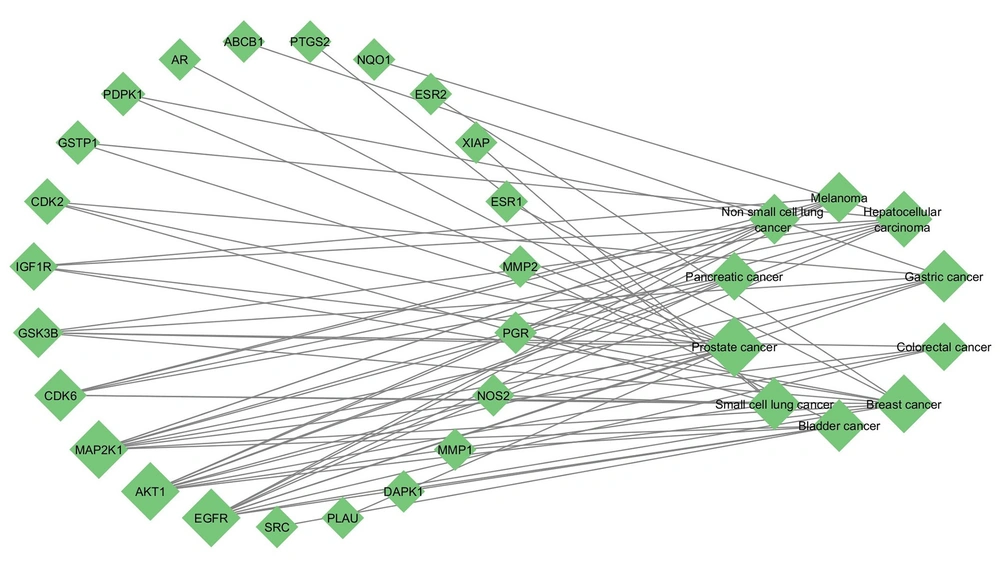
Additionally, enrichment analysis of the 37 intersecting targets within KEGG cancer pathways revealed the top 10 cancer pathways significantly associated with these targets (Table 1). The analysis showed that C. spinosa compounds can influence multiple components of pathways related to prostate cancer, breast cancer, bladder cancer, and hepatocellular carcinoma, among others — each involving at least two hub genes. A network view of these interactions is presented in Figure 5.
| Cancer | Adjusted P-Value | Odds Ratio | Targets |
|---|---|---|---|
| Prostate cancer | 1.22E-13 | 84.61472967 | GSK3B, AR, MAP2K1, PLAU, PDPK1, GSTP1, CDK2, AKT1, EGFR, IGF1R |
| Breast cancer | 1.78E-10 | 46.17624224 | GSK3B, MAP2K1, CDK6, AKT1, PGR, ESR1, EGFR, ESR2, IGF1R |
| Bladder cancer | 3.20E-09 | 110.2009217 | MAP2K1, SRC, DAPK1, MMP1, MMP2, EGFR |
| Hepatocellular carcinoma | 1.43E-08 | 34.14310345 | GSK3B, NQO1, MAP2K1, CDK6, GSTP1, AKT1, EGFR, IGF1R |
| Small cell lung cancer | 2.07E-07 | 44.73443361 | CDK6, NOS2, CDK2, XIAP, AKT1, PTGS2 |
| Melanoma | 1.52E-06 | 46.39925373 | MAP2K1, CDK6, AKT1, EGFR, IGF1R |
| Non-small cell lung cancer | 1.52E-06 | 46.39925373 | MAP2K1, CDK6, PDPK1, AKT1, EGFR |
| Gastric cancer | 1.91E-06 | 26.82607715 | GSK3B, MAP2K1, ABCB1, CDK2, AKT1, EGFR |
| Pancreatic cancer | 4.79E-05 | 33.48653199 | MAP2K1, CDK6, AKT1, EGFR |
| Colorectal cancer | 7.64E-05 | 29.38802661 | GSK3B, MAP2K1, AKT1, EGFR |
Network diagram of common Capparis spinosa targets and Kyoto encyclopedia of genes and genomes (KEGG) cancer pathways. The right circle of nodes represented the top enriched cancers, and the left nodes represented the common targets of the C. spinosa and cancer genes associated with each cancer.
4.5. Docking Results
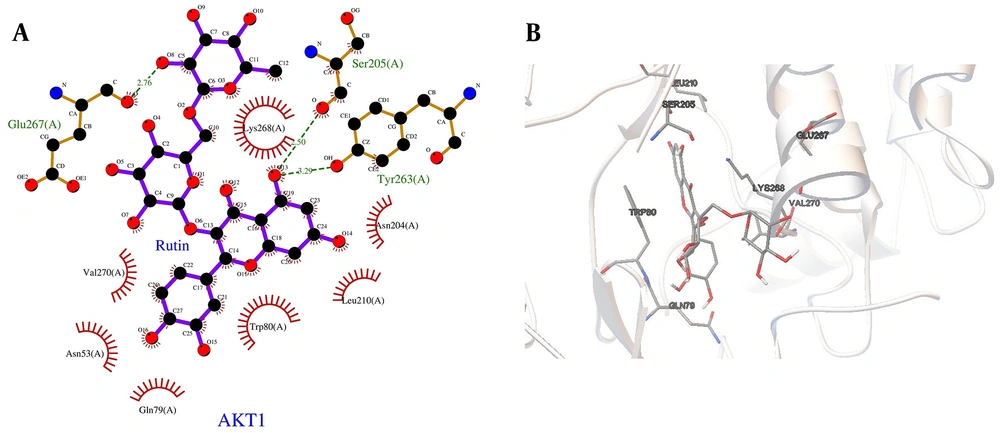
The identified cancer hubs — AKT1, EGFR, and SRC — were targeted with four compounds derived from Capparis species. AKT1 was associated with rutin, EGFR with flazin, kaempferol, and apigenin, and SRC with flazin. Our docking analysis showed that Capparis sp. compounds could interact with these cancer hub targets with strong binding affinities.
Rutin targeted AKT1 with a free binding energy of -5.18 kcal/mol and an inhibition constant (Ki) of 159.28 μM. Among the ligands of EGFR, apigenin exhibited the highest binding energy (-8.1 kcal/mol) and a Ki of 1.17 μM, followed by kaempferol (binding energy: -7.83 kcal/mol; Ki: 1.83 μM) and flazin (binding energy: -7.0 kcal/mol; Ki: 7.55 μM). Flazin also showed a strong interaction with SRC, with a binding energy of -8.08 kcal/mol and a Ki of 1.19 μM.
The docked complex of AKT1 with rutin, which demonstrated the best binding affinity, is shown in Figure 6 in both 2D and 3D views to illustrate the interacting groups and types of interactions (other complexes are presented in Appendix 4 in Supplementary File). Appendix 3 in Supplementary File provides a comprehensive list of all 31 compounds, along with their docking scores, inhibition constants, and key interactions with their respective targets, including AKT1, EGFR, and SRC.
5. Discussion
The present study provides a comprehensive analysis of the bioactive phytochemicals in C. spinosa and their potential therapeutic targets. The identification of 31 compounds with predicted targets offers a promising foundation for understanding the plant’s medicinal properties. Notably, the high abundance of compounds such as apigenin, kaempferol, and gallic acid suggests their significant roles in the plant’s pharmacological effects.
Gene ontology and enrichment analyses revealed that the identified targets are involved in crucial biological processes, including protein modification, phosphorylation, steroid metabolism, and the regulation of inflammatory and oxidative responses. These processes are fundamental to the plant’s capacity to exert antidiabetic, anticancer, anti-inflammatory, and neuroprotective effects. The involvement of these targets in key KEGG pathways further supports the potential of C. spinosa compounds in modulating disease-related mechanisms.
Traditionally, caper has been extensively used as a glucose-lowering herb in Iran and many other countries. Numerous studies have demonstrated its hypoglycemic and antidiabetic effects; however, the exact mechanisms remain unclear (7, 14). According to our pathway enrichment analysis, carbonic anhydrase (CA) activity in the nitrogen metabolism pathway, the FoxO signaling pathway, and anti-inflammatory/anti-oxidative targets may be associated with the observed antidiabetic effects.
Carbonic anhydrases are considered novel therapeutic targets for managing diabetes and its complications. The CA pathway plays a pivotal role in the regulation of glucose homeostasis, a key factor in diabetes management (15, 16). Inhibition of CA isoforms II and V has been shown to affect gluconeogenesis, a crucial process for hepatic glucose production (17). Specifically, CA V, located in the mitochondria, is essential for converting pyruvate to oxaloacetate — an integral step in both gluconeogenesis and lipogenesis (18). Inhibiting these isoforms has also been associated with weight loss, which is beneficial for individuals with type 2 diabetes mellitus (T2DM) and obesity, conditions that frequently coexist (19, 20).
We compared the CA inhibition activity of gallic acid — a compound found in C. spinose — with that of the clinically used CA inhibitor, acetazolamide. Gallic acid showed binding energies of -6.8 kcal/mol (Ki = 10.2 µM) for CA II and -7.1 kcal/mol (Ki = 6.5 µM) for CA IX, indicating moderate inhibitory activity. In comparison, acetazolamide demonstrated stronger binding affinities, with binding energies of -9.3 kcal/mol (Ki = 0.08 µM) for CA II and -9.5 kcal/mol (Ki = 0.05 µM) for CA IX. While gallic acid’s binding was weaker than that of acetazolamide, it still falls within the bioactive range and is comparable to other natural CA inhibitors, such as quercetin (-6.5 kcal/mol for CA II). These findings suggest that gallic acid may contribute to the antidiabetic and anticancer effects of C. spinosa by partially inhibiting CA activity, particularly in conditions where CA isoforms such as CA II and CA IX are overexpressed.
Among the Capparis phytochemicals, gallic acid and syringic acid were found to target both CA II and CA V, while seven other compounds were identified to target only CA II.
Furthermore, the FoxO signaling pathway is known to regulate genes involved in apoptosis, cell-cycle control, glucose metabolism, and oxidative stress resistance (21-23). Phosphorylation of FoxO proteins by Akt/protein kinase B (Akt/PKB), in response to insulin, leads to their nuclear export, thereby reducing the expression of genes that promote gluconeogenesis and enhancing insulin sensitivity (24). Conversely, during insulin resistance (IR), FoxO activity increases due to impaired PI3K/Akt signaling, contributing to elevated hepatic glucose production (25). In skeletal muscle, FoxO1 reduces glucose uptake and oxidation, promotes lipid uptake and oxidation, and increases muscle atrophy (26). Studies have shown that FoxO1 lowers pancreatic insulin production and secretion, and elevated FoxO1 activity in the hypothalamus increases the risk of developing type 2 diabetes (T2D) (27). Within the insulin signaling pathway, upstream targets of FoxO such as IGF1R, PDPK1, and AKT1 were associated with spinosin A, adenosine, and rutin, respectively.
Increased reactive oxygen species (ROS) and oxidative stress are also considered key contributors to the development of IR, T2D, and its complications. In diabetes, chronic hyperglycemia and mitochondrial dysfunction enhance ROS production, exacerbating oxidative stress (28). This oxidative burden negatively impacts various aspects of diabetes, including impaired β-cell function and IR, thereby disrupting glucose homeostasis (29). Antioxidants from C. spinosa exhibit protective effects against diabetes and its complications, particularly through β-cell protection and regeneration. These effects are attributed to its antioxidant phytochemicals, such as phenolic compounds, flavonoids, carotenoids, tocopherols, and terpenes (30).
Within the canonical insulin signaling pathway, PDPK1, AKT1, and GSK-3β were linked to C. spinosa compounds. Additionally, several negative regulators of insulin receptor signaling — such as PTPN1 (protein tyrosine phosphatase), JNK, and p38 — were identified as targets of C. spinosa phytochemicals. Specifically, apigenin targeted all three, kaempferol and isorhamnetin targeted JNK and p38, flazin targeted PTPN1 and p38, gentisic acid targeted PTPN1, capparine A targeted JNK3, and oroxylin targeted p38.
Capparis spinosa also holds promise as a therapeutic agent for managing diabetes-related complications. Mapping C. spinosa targets onto KEGG’s "AGE-RAGE signaling pathway in diabetic complications" revealed protective effectors relevant to diabetic nephropathy, cardiomyopathy, and non-alcoholic steatohepatitis (NASH). In this pathway, in addition to JNK3 and p38 MAP kinases, NOX4 is considered a major source of oxidative stress in diabetic kidney and vascular complications (31). In our study, NOX4 was identified as a target of kaempferol, apigenin, and astragalin.
These factors, along with matrix metalloproteinases (MMP1, 2, 7, 13), selectins (E and P), cyclooxygenase-2, and NLRP3, contribute to inflammatory processes that may be alleviated by C. spinosa phytochemicals.
Research has demonstrated that C. spinosa extracts possess neuroprotective properties. For instance, an aqueous extract of caper rich in rutin and quercetin was shown to attenuate cognitive impairment and reduce inflammation by modulating Alzheimer’s-related genes such as BACE1, APP, PSEN1, and PSEN2 (32). In our analysis, the Alzheimer’s disease pathway was significantly enriched by the predicted targets. β-Secretase (BACE1) plays a central role in the generation and modulation of Aβ peptides and is considered a key therapeutic target in Alzheimer’s disease (33). In this study, BACE1 was predicted to be targeted by kaempferol, apigenin, and ginkgetin among the C. spinosa -derived compounds.
Alpha-synuclein (α-syn) interacts with amyloid-beta (Aβ) and tau — two other hallmark proteins in AD — facilitating their aggregation (34). Gallic acid, one of the C. spinosa compounds, was associated with α-syn and has shown potential to inhibit its aggregation (35). Protein kinases such as glycogen synthase kinase 3 (GSK3β) and cyclin-dependent kinase 5 (CDK5) are implicated in the phosphorylation of tau, a process closely associated with neurodegeneration in AD (36). These kinases were targeted by kaempferol and apigenin in our results, supporting the potential of C. spinosa in reducing tau pathology and neuroinflammation.
Our findings are consistent with prior studies highlighting the anticancer potential of C. spinosa (37, 38). Notably, the KEGG pathways "Chemical carcinogenesis" and "Pathways in cancer" were significantly enriched based on our predicted targets (via EnrichR). Reactive oxygen species-mediated carcinogenesis can be mitigated by the antioxidant properties of C. spinosa phytochemicals. Chemical carcinogenesis, often mediated by cytochrome P450 enzymes, may be attenuated by targeting CYP1 with apigenin, kaempferol, and isorhamnetin. Apigenin also targets CYP19A1 (aromatase), a key enzyme in estrogen biosynthesis and a validated target in hormone-responsive cancers (39).
Moreover, receptor-mediated carcinogenesis may be influenced by C. spinosa compounds due to their potential interactions with androgen, progesterone, and estrogen receptors. These interactions were associated with apigenin, kaempferol, spinosin A, and β-sitosterol. Among the enriched pathways, CA pathways yielded the highest statistical significance. Carbonic anhydrases— particularly CA IX and CA XII — are frequently overexpressed in various tumors (40). Their role in pH regulation and modulation of the tumor microenvironment makes them valuable therapeutic targets (41). An acidic microenvironment promotes tumor invasion and angiogenesis (42). The CA IX, a major enzyme in this process, was predicted to be targeted by gallic acid, syringic acid, gentisic acid, and protocatechuic acid in our study.
The intersection of our predicted targets with cancer-associated genes underscores the anticancer potential of C. spinosa. To contextualize the docking performance of C. spinosa compounds, we compared their binding affinities with those of known reference inhibitors for AKT1, EGFR, and SRC:
AKT1: Rutin (C. spinosa compound) had a binding energy of -5.18 kcal/mol (Ki = 159.28 µM), which is lower than the reference inhibitor MK-2206 (-8.2 kcal/mol, Ki = 0.9 µM), yet still within the bioactive range.
EGFR: Apigenin and kaempferol demonstrated strong binding energies of -8.1 kcal/mol (Ki = 1.17 µM) and -7.83 kcal/mol (Ki = 1.83 µM), respectively. These values are slightly weaker than Gefitinib (-9.5 kcal/mol, Ki = 0.02 µM) but comparable to other natural EGFR inhibitors.
SRC: Flazin showed a binding energy of -8.08 kcal/mol (Ki = 1.19 µM), moderate compared to Dasatinib (-10.2 kcal/mol, Ki = 0.03 µM), but in line with quercetin (-7.5 kcal/mol), a natural SRC inhibitor.
Although the binding affinities of C. spinosa compounds were generally weaker than those of pharmaceutical reference drugs, they remain within the range of bioactive compounds. These results support the multi-target therapeutic potential of C. spinosa in neurodegenerative and cancer-related conditions.
While this study provides valuable insights into the potential pharmacological mechanisms of C. spinosa using network pharmacology and molecular docking, it has several limitations.
First, the study relies entirely on computational predictions without experimental validation. Although these methods are widely accepted in drug discovery, in vitro and in vivo experiments are necessary to confirm the predicted interactions and pathway involvement. In future research, we plan to conduct in vitro assays (e.g., enzyme inhibition, cell-based studies) and in vivo experiments using animal models to evaluate the effects of C. spinosa compounds on diabetes- and cancer-related pathways.
Second, the selection of active compounds was based on OB and DL criteria. While these filters improve relevance, they may inadvertently exclude some bioactive agents or fail to account for the effects of metabolites and compound synergy. Moreover, the study did not assess ADMET (absorption, distribution, metabolism, excretion, and toxicity) properties, which are critical for the clinical translation of any therapeutic compound.
Finally, although the multi-target nature of C. spinosa compounds may offer therapeutic advantages, it also raises concerns about potential off-target effects and complex dose-response relationships — factors not addressed in this study. Future research should therefore include experimental validation, improved docking methods, and comprehensive ADMET profiling to more thoroughly evaluate the therapeutic potential of C. spinosa.
5.1. Conclusions
In this study, we employed network pharmacology and molecular docking to investigate the potential pharmacological mechanisms of C. spinosa in the treatment of diseases such as diabetes and cancer. We identified 31 active compounds in C. spinosa and predicted their interactions with 183 protein targets, including key hubs such as AKT1, EGFR, and SRC. Compounds such as apigenin, kaempferol, and gallic acid showed strong binding affinities to these targets, suggesting their potential role in modulating pathways involved in diabetes, cancer, and inflammation.
Although the binding affinities of C. spinosa compounds were generally lower than those of reference drugs, they were still comparable to other known natural bioactive agents, supporting their potential as multi-target therapeutic compounds. Overall, this study lays a foundational understanding of the complex mechanisms underlying C. spinosa activity and highlights its promise as a versatile medicinal plant for the treatment of multiple diseases.