1. Background
Myocardial infarction (MI) is the death of myocardial cells because of prolonged ischemia (1). Thrombolytic therapy is an important form of treatment for acute MI (2). In this regard, different fibrinolytic agents and plasminogen activators (PAs) have been developed (2). Streptokinase and urokinase (as the first generation agents); alteplase (as the second generation agent); and reteplase, tenecteplase, lanatoplase, and staphylokinase (as the third generation agents) are three types of PAs in the market (3). It has been shown that immediate intravenous infusion of alteplase is associated with an improved mortality compared with intravenous administration of streptokinase or the combination of streptokinase and t-PA (2).
Tissue plasminogen activatoris a 527 amino acids glycoprotein with a molecular weight of 67 kDa, which causes conversion of plasminogen to plasmin in the presence of fibrin. The protein molecule contains five structural domains: finger domain (F), a growth factor domain (EGF), the N-terminal region, and two kringle 1 (K1) and kringle 2 (K2) domains. Kringle 2 domain is the serine protease domain with the catalytic site at the C terminus. Binding of both finger and kringle 2 domains to fibrin accelerates the activation of t-PA on plasminogen (4).
However, full t-PA has several disadvantages such as the rapid clearance from plasma by the liver as the structural elements on first three N-terminal domains are recognized by certain hepatic receptors (5-7), and difficult prokaryotic production and refolding process (8). Therefore, smaller active molecules such as reteplase and lanatoplase have been synthesized and are commercially available (3). Reteplase is a mutant version of t-PA with prolonged half-life in which the F, EGF, and K1 region of the wild-type t-PA molecule have been deleted. Finger domain is the responsible domain for fibrin affinity. Therefore, reteplase has weaker affinity for fibrin and causes more fibrinogen depletion than full length forms (2, 9). Furthermore, reteplase can be used within 3 hours of stroke, while streptokinase is not indicated for treatment of stroke (10).
2. Objectives
This study aimed at the expression of reteplase in optimum condition. In this study, the reteplase gene was cloned and expressed in Escherichia coli TOP10 as a suitable host cell, and its expression was optimized.
3. Materials and Methods
3.1. Materials
The pET15b cloning and expression vector were obtained from Novagen Co., USA. The pBAD/gIIIA plasmid and bacterial strains E. coli TOP10 and E. coliDH5α were purchased from Pasteur Institute and Cinnagen, Iran, respectively. Luria-Bertani (LB) media was prepared according to the guidelines recommended by Sambrook and Russell (11). Screening was performed based on antibiotic resistance tested on ampicillin-containing LB agar plates (for screening the colonies based on antibiotic resistance, 100 µg/mL) obtained from Sigma, Germany (12), plasmid minipreparation kit and extraction quick gel kit were purchased from Fermentas Co., Poland, and QIAgen, Germany, respectively.
3.2. Methods
3.2.1. Transformation of E. coli DH5α With Recombinant pET15b Plasmids
One hundred microliter of competent E. coli DH5α were transformed with recombinant pET15b/reteplase plasmid using CaCl2 and heat shock transformation method (39°C, 1 minute). The transformed cells were spread on LB agar plates containing 100 mg/mL of ampicillin. After overnight incubation at 37°C, some colonies were selected for plasmid minipreparation using kit method. Plasmids were detected in 0.8% agarose gel electrophoreses. The plasmid pET15b/reteplase was digested by NcoΙ and BamHΙ restriction enzymes to obtain reteplase cDNA insert. In the meantime, pBADgIIIA vector was digested by NcoΙ and BglΙΙ enzymes for 45 minutes. In order to prevent the attachment of linear plasmids, alkaline phosphatase was added to the mixture and put at 65°C for 10 minutes.
These digestions were separated in gel electrophoresis using extraction quick gel kit (QIA, Germany). Then these two products with loading buffer sample and DNA leader and a sample of λ marker were electrophoresed, and the amount of DNA in samples was determined according to the λ marker. Vector and insert (molar ratio of 3:1, vector to insert) were ligated by T4 DNA ligase. Next, E. coli TOP10 was transformed using heat shock method (39°C, 1 minute) and spread on ampicillin (100 µg/mL)-containing LB agar plates and incubated overnight at 37°C. Afterward, the obtained recombinant pBAD/gIIIA plasmids (using alkaline lysis), were sequenced by Gene Fanavaran company, using the Analyzer Genetic Device and Capillary Base. Finally, digestion was performed by EcoRI restriction enzyme.
3.2.2. Expression
One colony of E. coli TOP10 containing recombinant plasmid was cultured in 5 mL of LB medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) at 37°C, shaking at 200 rpm overnight. Then, 25 µL of this culture was inoculated into 25 mL of LB medium containing ampicillin (100 µM) at 37°C. About 2 hours after reaching OD600 to 0.4-0.6, the sample was divided into 4 parts. Then, L-arabinose was added in concentration of 0.2, 0.02, 0.002, and 0.0002% (prepared by serial dilution) to each part. The final OD600 of inoculums in any experiment was read after 4 hours and the samples were centrifuged (5000 × rpm), at 4°C for 10 minutes. These pellets were stored at -20°C (13, 14) and one that had the most OD600 was used for the other stages.
3.2.3. Extraction of Reteplase From Periplasmic Space
Pellets from the best concentration of L-arabinose were resuspended in hypertonic buffer and then edetic acid (EDTA) 500mM was added to the sample and put on ice for 10 minutes and shook. The sample finally was centrifuged for 20 minutes at 4°C (8000 × rpm) and the supernatant was discharged, the pellet was resuspended in MgSO4 solution and centrifuged for 20 minutes at 4°C (8000 × rpm). The supernatant as the periplasmic protein was stored at 4°C. This solution was used for SDS-PAGE (Sodium dodecyl sulfate Polyacrylamide gel electrophoresis)analysis and purification using nickel resin affinity chromatography (15).
3.2.4. Preparation of Inclusion Bodies
Some pellets were resuspended in 150 mL of 0.1 M Tris and 20 mM EDTA and homogenized using a shearing rod, Micro Smash (Tomy, Japan). A solution of lysozyme (0.25 mg/mL) was added to the samples and incubated for 30 minutes on ice. Then, centrifugation was carried out for 50 minutes at 4°C (13000 × rpm). The pellets were resuspended in 90 mL of 0.1 M Tris, 20 mM EDTA and 2.5% v/v Triton X-100 and homogenized again. Next, the samples were centrifuged, and resuspended in 90 mL of 0.1 M Tris, 20 mM EDTA and 0.5% v/v Triton X-100 solution and homogenized. The samples were then centrifuged for 30 minutes at 4°C (13000 × rpm) and the pellets were resuspended in 75 mL of 0.1 M of Tris and 20 mM EDTA (16, 17). The prepared inclusion bodies were stored at -20°C.
After preparation of inclusion bodies, proteins were extracted by resuspension in Tris 25 mM, EDTA 10 mM, containing 1% Triton X-100 and guanidine HCl 6 M (18). The reducing agent and buffer components were separated by dialysis (pH = 7) at 4°C. The solubilized samples were incubated in a mixture of Tris 0.1 mol/L (pH = 8.5), guanidine hydrochloride 6 mol/L, EDTA 2 mmol/L, and -mercaptoethanol 1%. Subsequently, the pH of the solution was adjusted to 7 with HCl (12 N). Refolding of the protein was done by dilution with 0.1 mol/L Tris (pH = 10.5), 0.5 M L-argenine, 1 mM EDTA, 6 Mguanidine hydrochloride, 1 mM reduced glutathione, 0.1 mM oxidized glutathione, and 1 mg/mL bovine serum albumin. Then, the samples were incubated for 24 hours at 20°C in 180 rpm. The reducing agent and buffer components were separated by dialysis of the protein samples using 0.1 M Tris and 1mMEDTA (pH = 8), and this stage was repeated three times for 1 hour and final repetition was performed for 24 hours.
3.2.5. SDS-PAGE Analysis
The inclusion bodies were dissolved in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 Mm KH2PO4, 0.05% Tween 20, pH = 7.4) and these samples, periplasmic solution and refolded protein were boiled for 5 minutes at 70°C and electrophoresed on a 12% (v/v) SDS-PAGE analysis. Subsequently, the gel was stained with Coomassie Brilliant G250.
3.2.6. Purification of Reteplase Using Nickel Resin Affinity Chromatography
In this project, Ni-NTA His-Bind Resin was used for purification of the protein with poly-His sequence. Ni-NTA His-Bind buffer was added to the pellets of refolded protein and then lysozyme 10 µg/mL was added to the samples, and the mixtures were vortexed and centrifuged at 15000 rpm for 10 minutes. The supernatant (periplasmic protein) was mixed with resin and shook for 30 minutes on ice. The samples were then centrifuged at 15000 rpm for 10 seconds, and the supernatant was discharged. Afterward, Ni-NTA His-wash buffer was added to the samples and centrifuged at 15000 rpm for 10 seconds and this stage was repeated. The pellets were washed with Ni-NTA His-elute buffer and the elutions were collected.
3.2.7. Measurement of the Activity of Reteplase
The detection of plasminogen activation by reteplase was done by Chromogenic Activity Assay Kit. Assay diluention (50 µL), plasminogen (10 µL), plasmin substrate (20 µL), and 20 µL oft-PA standard reteplase, the samples and water as blank were added to the supplied 96-well plate. The plate was incubated at 37°C in a humid incubator and the absorbance was measured at 405 nm for each sample.
3.2.8. Optimization of Reteplase Expression
In this study, the effect of temperature (25°C, 30°C, and 37°C), concentration of inducer (0.2%, 0.02%, 0.002%, and 0.0002%) and the time of incubation (1, 2, 3, and 4 hours) was checked using dot blot technique. The pellets of the best concentration of L-arabinose were put on ice, and the lysis buffer was added and resuspended. The suspension was frozen and thawed three times, put on nitrocellulose membrane, and dried at room temperature for 20 minutes. Afterward, the blocking solution (nonfat skim milk 5%) was seeded to the nitrocellulose membrane and shook for 1 hour. Primary antibody (anti-His) was then added to membrane and shook for 1.5 hours again. Next, the secondary antibody (anti-IgG conjugated with HRP) was added to the membrane and shook for another 1.5 hours. Finally, OPD solution (HRP substrate) was added to wash nitrocellulose membrane, and the spots were detected. The intensity of each spot was determined using Photo Capture software, and the results were assessed using design expert software.
4. Results
4.1. Extraction of Recombinant pET15b Plasmids and pBAD/gIII From E. coliDH5α
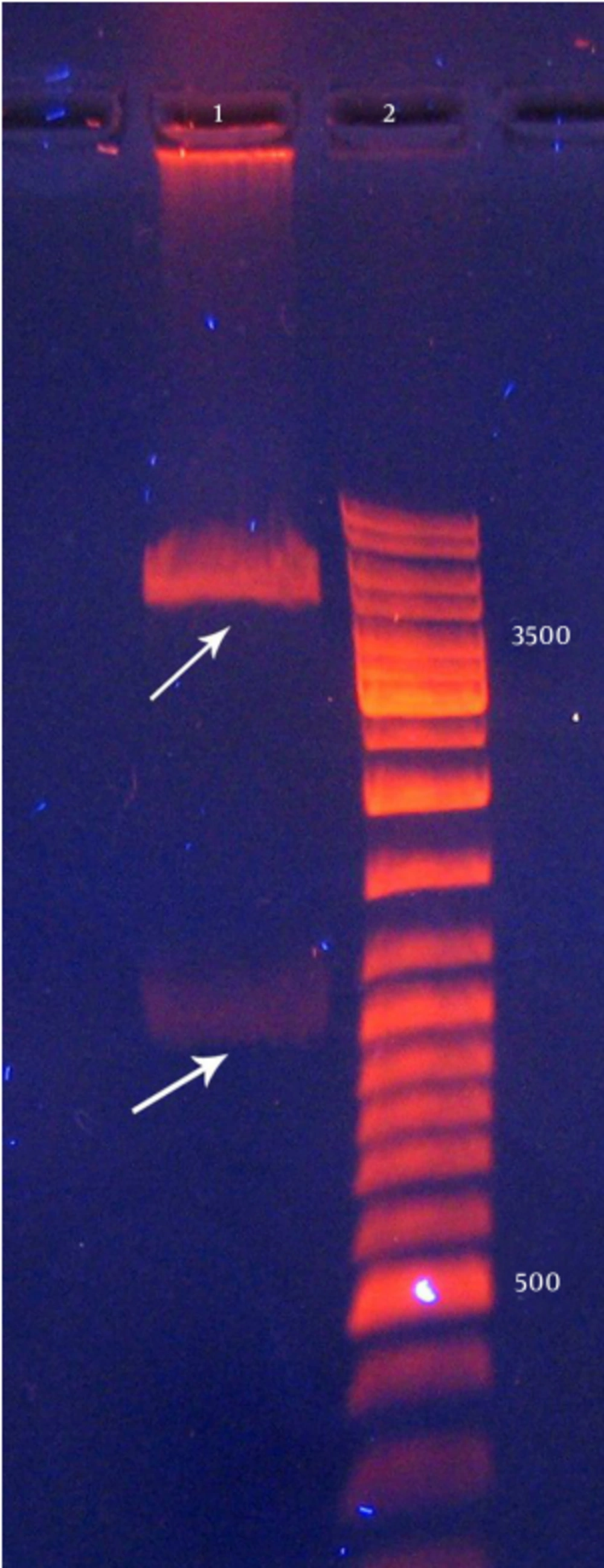
Extraction of recombinant pET15b plasmids and pBAD/gIIIA from E. coliDH5α was performed using alkaline lysis. Then, the sample was electrophoresed (Figure 1) and the presence of these plasmids was confirmed.
4.2. Digestion of Recombinant pET15b Plasmids and pBAD/gIII
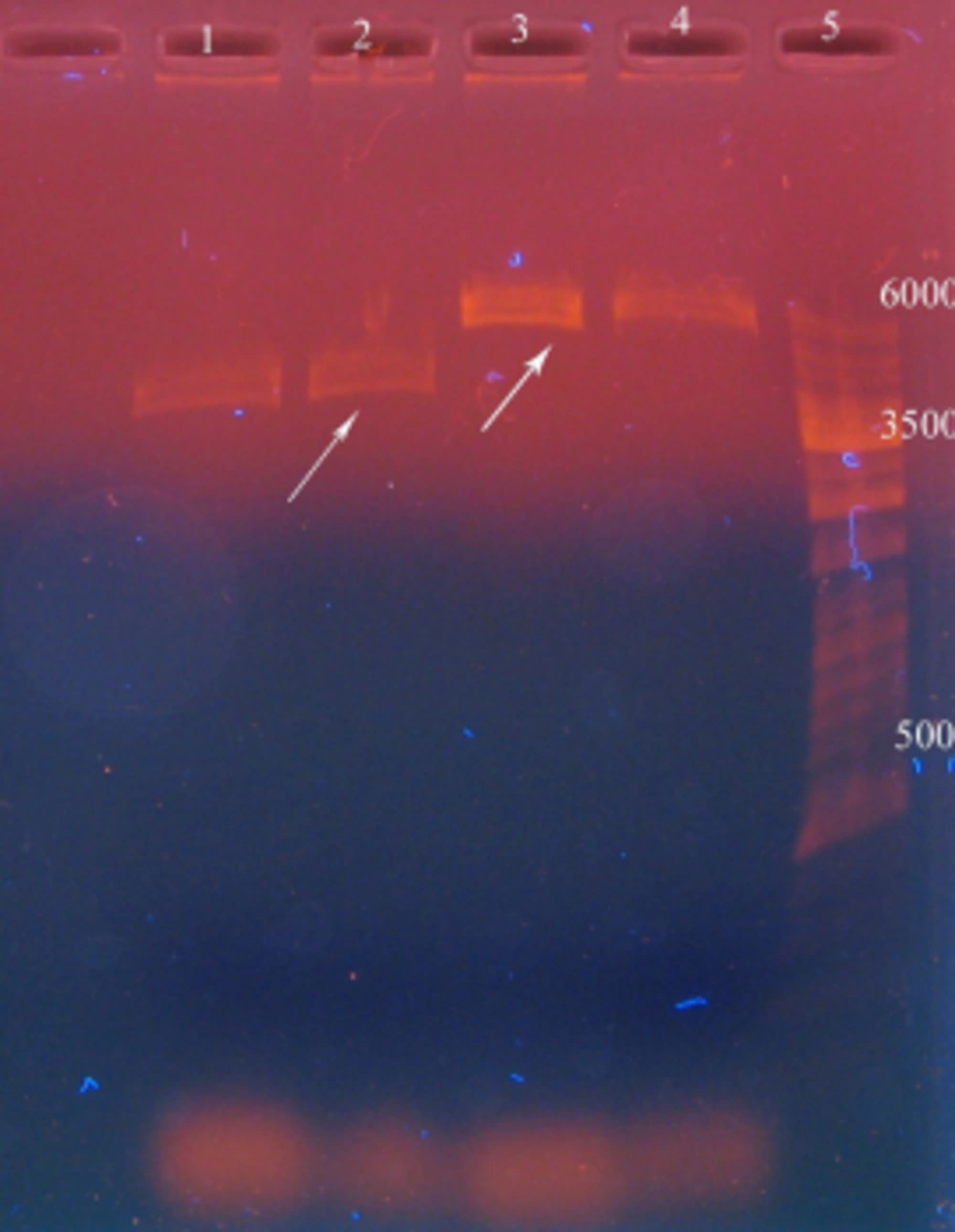
After the digestion of recombinant pET15b plasmids by BamHI and pBAD/gIIIA by NcoI and BglII, reteplase (1128 nucleotides) and linear pBAD/gIIIA (4145 nucleotides) were detected on agarose gel (Figure 2).
4.3. Quantification of the DNA using λ Marker
In this stage, the size of the insert and linear pBAD/gIIIA was confirmed using λ marker and unknown bands were compared with λ marker band and the bands were determined as 2.32 ng for the insert and 8.32 ng for pBAD/gIIIA.
4.4. Cloning of Reteplase
Recombinant pBAD/gІІIA plasmid containing reteplase was digested by EcoR1 restriction enzyme and three bands were observed (472, 520, and 3153 bp) and it showed the correct orientation of the insert in the recombinant pBAD/gІІI plasmid. As shown in Figure 1, the recombinant plasmids contained the correctly oriented insert. Sequencing of this plasmid also confirmed the presence of reteplase cDNA in the pBAD/gІІI plasmid. The insert (1128 bp) was separated using the HindIII and NcoI restriction enzymes.
4.5. Expression of Reteplase
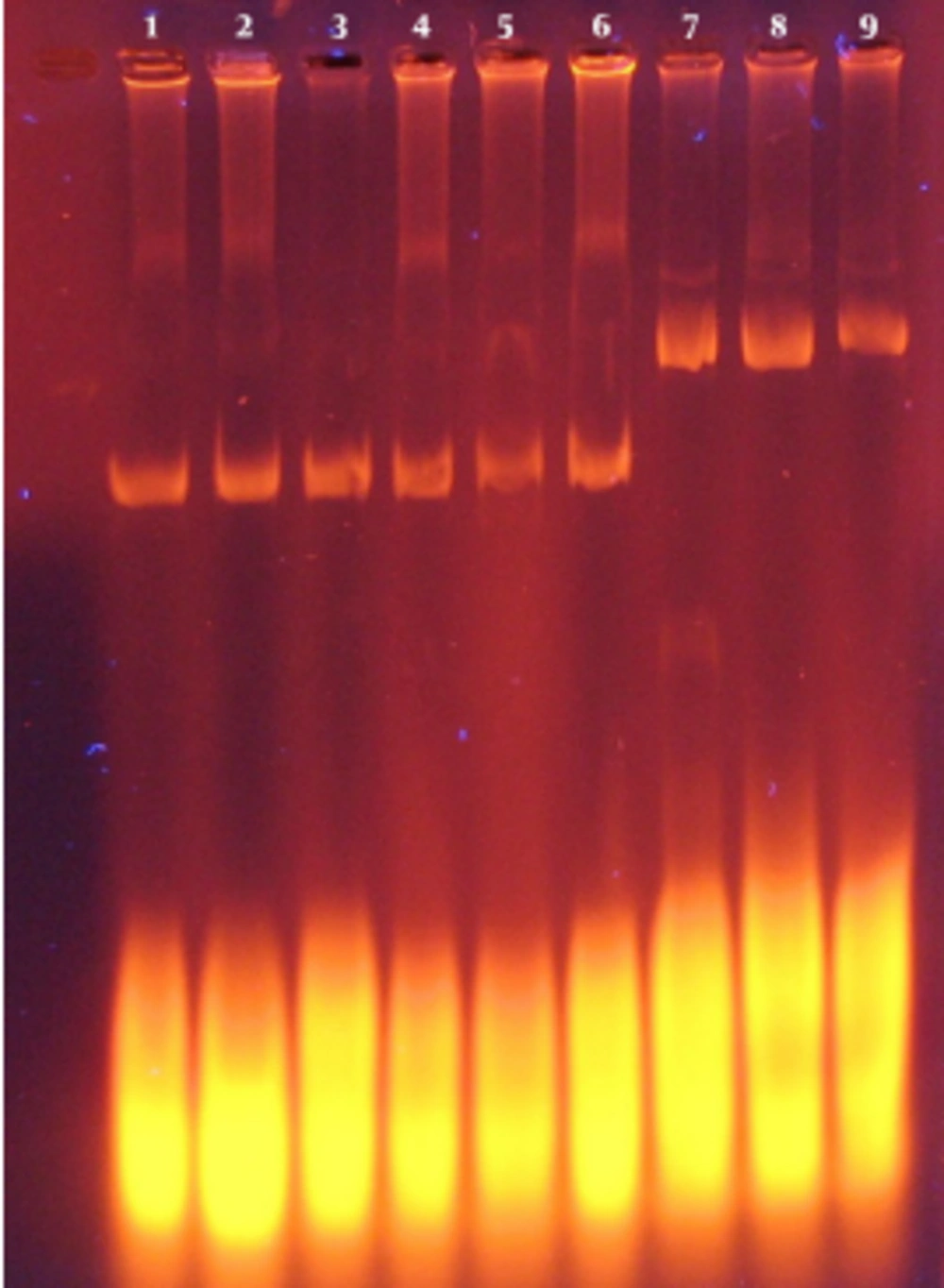
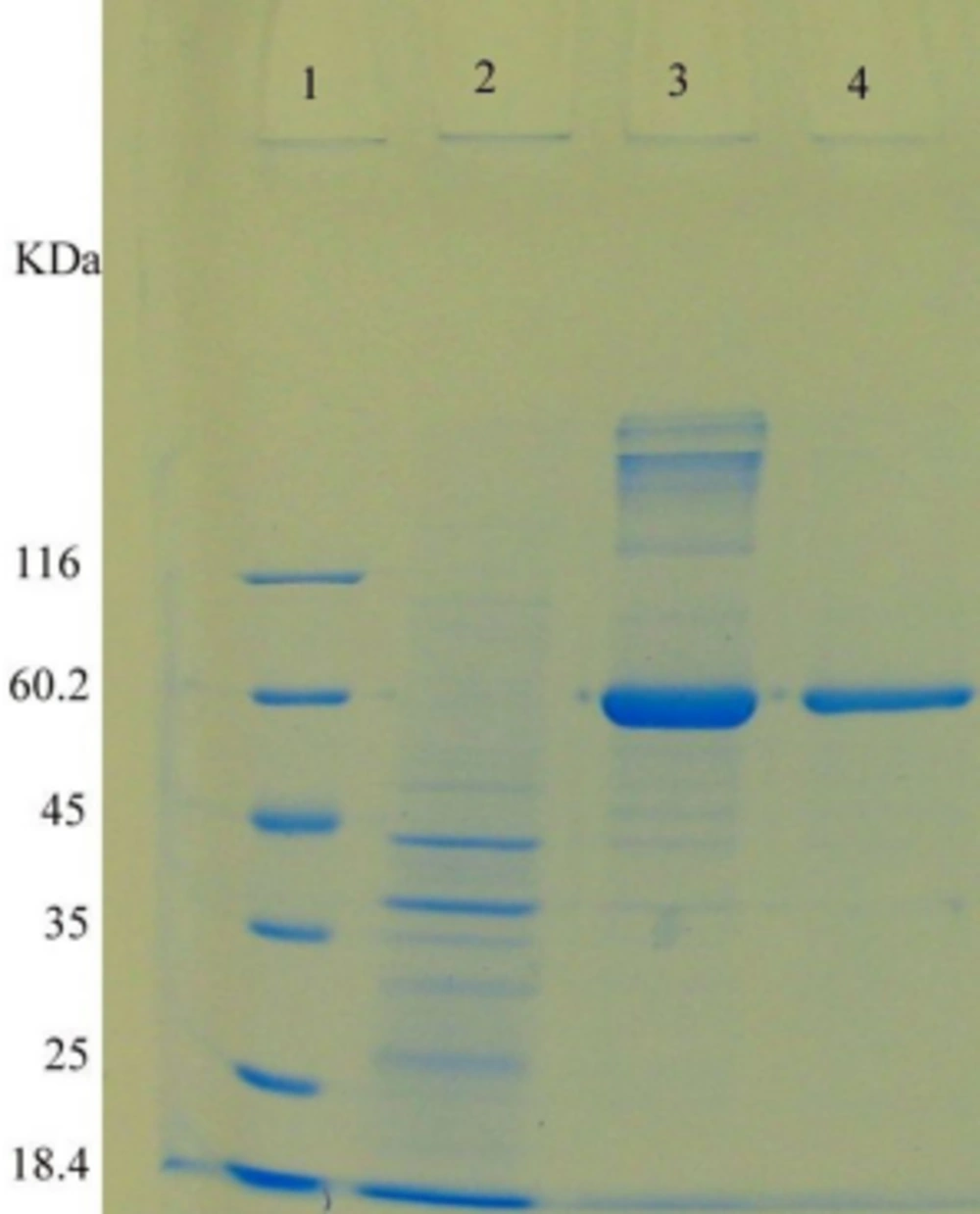
After the transformation of E. coli TOP10 with pBAD/gІІIA plasmid containing reteplase gene, the effect of L-arabinose (0.2, 0.02, 0.002 and 0.0002%) on protein expression was evaluated (2 hours incubation at 37°C). The final OD600 was determined as 1.3. The expressed proteins were electrophoresed using SDS-PAGE (Figures 2 and 3). Soluble proteins were isolated from the periplasmic space. A protein with an estimated size of ∼ 39 kDa was observed in SDS-PAGE (Figures 4, 5).
Lane 1, Standard molecular weight marker; lanes 2, 3, 5, 7, and 9, periplasmic sample of recombinant pBAD/gІІІA plasmids containing reteplase induced with 0.0002 % L-arabinose for 2 hours; lane 4, 6, 8, 8, and 10, inclusion body of recombinant pBAD/gІІІA plasmids containing reteplase induced with 0.0002 % L-arabinose for 2 hours.
4.6. Purification of the Refolded Protein
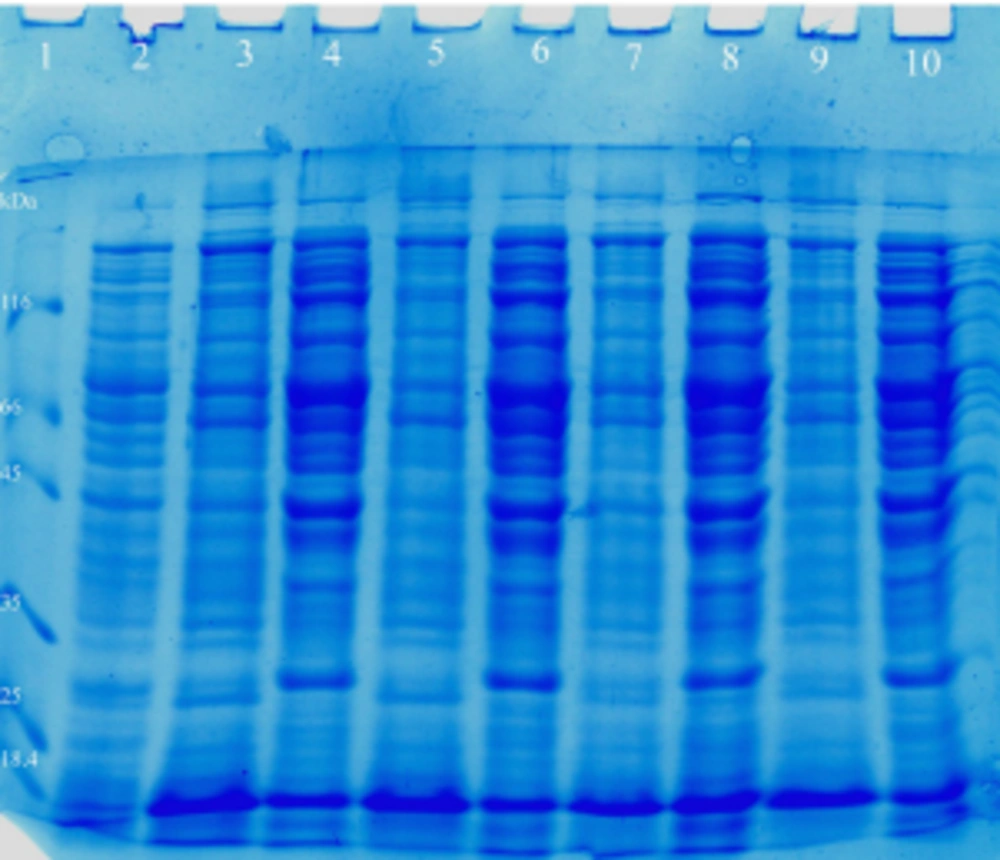
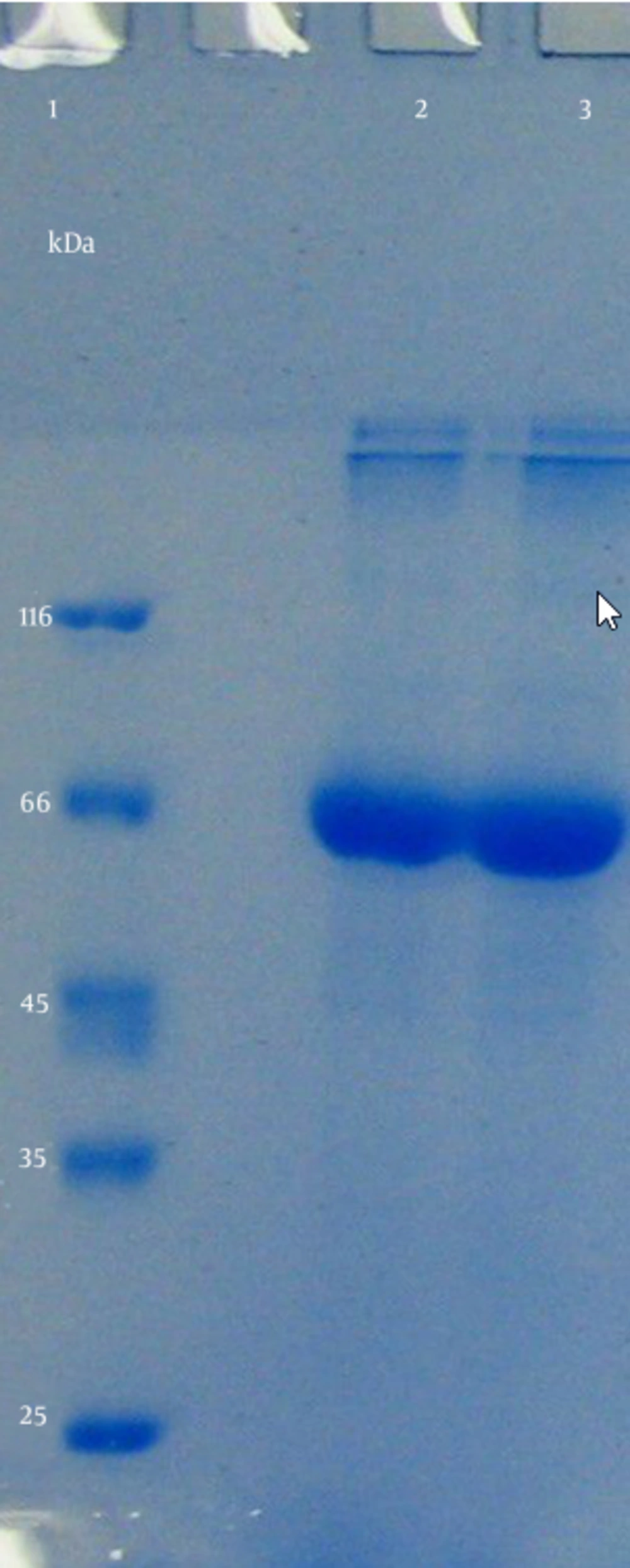
After the purification of periplasmic reteplase using nickel resin affinity chromatography the sample was analyzed using SDS-PAGE electrophoresis, and one band was observed at about 66 kDa (Figure 6).
4.7. Activity of Reteplase
The activity of the obtained enzyme was analyzed using standard t-PA enzyme activity kit. The change in the absorbance of reaction solution at 405 nm was directly proportional to the t-PA enzymatic activity. The concentration of t-PA standard was 1 unit equivalent to12 µg/mL. The enzymatic activity of samples was measured as 0.8 units compared with the standards.
4.8. Optimizing the Expression of Reteplase
The intensity of each spot was determined using Photo Capture software and finally the results were assessed using Design Expert software indicated that the highest intensity of mixture the conditions is related to 37°C temperature, at 0.0002% L-arabinose, and for 2 hours incubation (64.542 sqrt). Table 1 presents these results. Increasing the inducer concentration and incubation time doesn’t increase the amount of protein expression. Furthermore, according to the results from software, the best temperature for optimum expression is 37°C and even with high concentrations of inducer, a low temperature reduces the amount of protein expression.
| Number | Temperature, ˚C | Inducer Concentration, µM | Time of Incubation, h | Intensity, Squrt | Desirability |
|---|---|---|---|---|---|
| 1 | 37 | 0.0002 | 2 | 64.542 | 0.782 |
| 2 | 37 | 0.0002 | 3 | 63.382 | 0.765 |
| 3 | 30 | 0.0002 | 2 | 61.151 | 0.733 |
| 4 | 37 | 0.0002 | 1 | 60.708 | 0.727 |
| 5 | 25 | 0.0002 | 3 | 59.099 | 0.704 |
| 6 | 37 | 0.002 | 2 | 56.784 | 0.670 |
| 7 | 30 | 0.0002 | 1 | 56.683 | 0.669 |
| 8 | 30 | 0.02 | 2 | 56.331 | 0.664 |
| 9 | 37 | 0.2 | 1 | 56.328 | 0.664 |
| 10 | 37 | 0.02 | 2 | 54.492 | 0.637 |
| 11 | 30 | 0.2 | 1 | 54.448 | 0.636 |
| 12 | 37 | 0.2 | 2 | 54.357 | 0.635 |
| 13 | 30 | 0.2 | 2 | 53.112 | 0.617 |
| 14 | 25 | 0.02 | 2 | 52.778 | 0.612 |
| 15 | 25 | 0.0002 | 2 | 52.726 | 0.612 |
| 16 | 37 | 0.0002 | 4 | 52.464 | 0.608 |
| 17 | 30 | 0.002 | 2 | 52.323 | 0.606 |
| 18 | 25 | 0.002 | 3 | 51.554 | 0.595 |
| 19 | 25 | 0.0002 | 4 | 51.464 | 0.593 |
| 20 | 30 | 0.0002 | 3 | 50.716 | 0.585 |
5. Discussion
Reteplase is an important drug for the treatment of myocardial infarction and brain stroke. Several studies have demonstrated that treatment with t-PA resulted in a reduced mortality of 1.2% when given within 70 minutes of the onset of chest pain compared with 8.7% when given between 70 minutes and 3 hours after the onset of pain (19-22). Therefore, the present research was conducted to produce recombinant reteplase in E. coli TOP10 in optimum condition. Induction of proteins using pBAD/gІІIA system has been reported by many studies (23, 24), although none of them have studied the expression of reteplase in this system. The most important advantage of pBAD/gІІIA vector is the presence of a signal sequence at the N-terminal of reteplase gene enabling it to be secreted into the periplasmic space, and this provides a better environment for proper folding of this enzyme (23).
In our study, low amount of the expressed reteplase was detected in periplasmic space while most of it was present inside the cell as inclusion bodies. This might be due to high expression of proteins disturbing the export mechanisms of cells for exporting recombinant proteins to the periplasmic space. Similar results have been reported by other investigators that produced proteins such as leucine-specific binding protein of Escherichia coli and reteplase in periplasmic space (12, 13, 25). The inclusion bodies have two main advantages of being mostly composed of recombinant proteins and easy isolation from the cell debris (12, 26). However, inclusion bodies are insoluble and mainly inactive (12, 27, 28); thus the main problem in purification process of inclusion bodies is optimization of the refolding and renaturation states by prevention of the formation of inactive aggregates.
Different strategies have been used for refolding the protein. In this study, we used arginine and oxidizing/reducing glutathione. The process of refolding reteplase may result in dimerization of the protein or changing its disulfide bands so that its molecular weight shows higher values in the gel although it is active. The molecular weight of the commercially reteplase is 39 kDa and it has more activity than our product. Therefore, it seems that changing the refolding protocol is necessary. The activity of our product is lower than that of the standard control. One possible reason for this is the presence of His-tag in the sequence of the gene that facilitates the purification of reteplase using affinity chromatography with nickel in stationary phase.
Qiu et al. Lee have also produced tissue plasminogen activators and measured their activities (26, 29). However, the unit activity of reteplase is presented differently as compared to other types of t-PA; therefore, the results of these studies cannot be compared with our work. One of the most important advantages of the periplasmic secretion of reteplase is its simpler purification. The presence of contaminations such as endotoxins in this space is less, and the quality of the product will increase (30, 31). One of the most important factors that influence the expression of recombinant proteins is the usage codon phenomenon. The frequencies of codons in eukaryotic and prokaryotic organisms are different; this difference results in the inhibition of expression, premature termination of translation, plasmid instability, and production of inactive proteins (32, 33). There are 20 rare codons in the reteplase gene and this is an important factor interfering with protein expression in E. coli (12). It can be resolved by inducing mutations in genes or codon substitutions.
In this project, we used araBAD promoter, which is weaker than T7 promoter but can be precisely controlled using different concentrations of L-arabinose and can produce soluble or insoluble proteins (34, 35). An important aspect of our study is the optimized expression of the reteplase. In conclusion, in this study, the expression of reteplase in Escherichia coli TOP10 was performed in an optimum condition. In future studies, it is suggested that, removing the His-tag from the sequence of reteplase be performed.