1. Background
Sepsis a symptomatic bacteremia is a frequent condition in cancer patients with a significant morbidity and mortality worldwide (1). This condition is a medical crisis that needs broad-spectrum antibiotic treatment. The gold standard for diagnosis of bacteremia is determination of etiology from blood culture (2). Among cancer patients, blood culture positive rates ranging from 2.6% to 28.4% has been reported from various hospitals in Iran (3-5).
Among cancer patients, as bacteremia is a life-threatening emergency, the information of epidemiological pattern of pathogens in a given district helps to inform the choice of antibiotics. Prevalence of bacterial isolates is influenced by geographic location and different times. Some bacteria commonly isolated include Escherichia coli, Klebsiella pneumonia, Enterobacter species, Pseudomonas aeruginosa and Staphylococcus aureus (6, 7).
2. Objectives
This study examined bacteremia in cancer patients from two centers regarding isolates and spectrum of antibiotic resistance pattern.
3. Patients and Methods
This was a prospective experimental investigation performed in Cancer Institute, Imam Khomeini Hospital, Tehran university of medical sciences, Tehran and Karaj Cities, Iran. All hospitalized patients with an underlying cancer with clinically significant Blood Stream Infection (BSI) were eligible for the study. After blood collection, samples were inoculated into blood culture bottles and incubated overnight at 37°C. Isolation and identification of the bacteria were performed by conventional microbiological techniques (8). This study was approved by the local ethics committee of University, which was held in 2012. In vitro antibiotic resistance pattern of the isolates was determined by the Clinical and Laboratory Standards Institute (CLSI) guidelines (9).
3.1. DNA Extraction
Bacterial colonies were obtained from clinical isolates and resuspended in saline and preceded for DNA extraction. Genomic DNA was isolated by DNA extraction kit (Roche, Mannheim, Germany). Bacillus cereus strain CF6, groEL, Escherichia coli strain K-12, phoA, Staphylococcus aureus strain MA042, femA and Staphylococcus aureus strain ATCC 43300 were used as a positive control for mecA and used other related strains as negative controls.
PCR amplification (Eppendorf, Germany) of target DNA was performed in a total volume of 50 μL. The reaction mixture contained 5 μL 10 × amplification buffer [500 mM KCl, 100 mM Tris/HCl (pH 8.5), 1.0% Triton X-100], 1 μL 25 mM MgCl2, 0.6 μL each of 2.5 mM dNTPs (Fermentas, GmbH, Germany), 1 μL forward and reverse primers for all genes (20 ng/μL), 0.4 μL Taq DNA polymerase (5 U/μL) and 200 pg extracted DNA. PCR conditions were initial denaturation at 94°C for 4 minutes, followed by 32 cycles of 94°C for 1 minute, 52.5°C for 1 minute and 72°C for 1 minute, with a final extension at 72°C for 5 minutes. Multiplex PCR was performed by simultaneous addition of primer pairs for phoA, femA, mecA and groEL in the same reaction mixture. Primer sequences used in this study are shown in Table 1. We designed and set up all primers by AlellID 6 software (manufactured by Premire, Biosoft International Palo Alto, California, USA.). Amplified products were separated by agarose (1%) gel electrophoresis in 0.5 × TBE, stained by ethidium bromide.
| Gene | Sequence 5’ → 3’ |
|---|---|
| phoA | F: GCACGTAATTATGCCGAA; R: TCAGCGCATAGTGAGTGTAT |
| femA | F: CATTGCAGCTTGCTTACTT; R: TCACGCTCTTCATTTAGTTCT |
| mecA | F: GATCATAGCGTCATTATTCCA; R: TAGTTCTTTAGCGATTGCTTTA |
| groEL | F: AAAGCTGTAGTTGCTGCAGTA; R: CAAATCCAGGAGCTTTAACA |
4. Results
During the study (April 2013 to October 2013), 68 blood culture bottles were received from patients with cancer, from which 12 (17.65%) samples had positive results. The distribution of pathogens obtained from the blood culture bottles of cancer patients is shown in Table 2. The most common bacterial pathogen isolated was E. coli, accounting for 5 (33.33%) samples. While each of S. aureus, B. cereus and K. pneumonia accounted for 3 (20%) samples.
| Antibiotic | Bacterial Number, % | ||||
|---|---|---|---|---|---|
| E. coli (5) | K. pneumonia (3) | S. aureus (3) | B. cereus (3) | Micrococcus spp. (1) | |
| Penicillin | 100 | 100 | 100 | 100 | 100 |
| Ampicillin | 100 | 100 | 0.0 | 0.0 | 0.0 |
| Gentamicin | 100 | 100 | 0.0 | 0.0 | 0.0 |
| Cefoxitin | 100 | 100 | 25 | 75 | 75 |
| Trimethoprim sulfamethoxazole | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Chloramphenicol | 25 | 25 | ND | ND | ND |
| Nitrofurantoin | 0.0 | 25 | ND | ND | ND |
| Tetracycline | 100 | 100 | ND | 100 | ND |
| Rifampin | ND | ND | 25 | 100 | 0.0 |
| Erythromycin | 75 | 75 | 50 | 100 | 0.0 |
| Mupirocin | ND | ND | 75 | 0.0 | 100 |
| Teicoplanin | ND | ND | 0.0 | 0.0 | 0.0 |
a Abbreviation: ND, Not done.
E. coli isolates showed maximum resistant to Penicillin, ampicillin, gentamicin, Cefoxitin, trimethoprim sulfamethoxazole and tetracycline. Similarly, S. aureus was 100% resistant to gentamicin and sulfamethoxazole (Table 2).
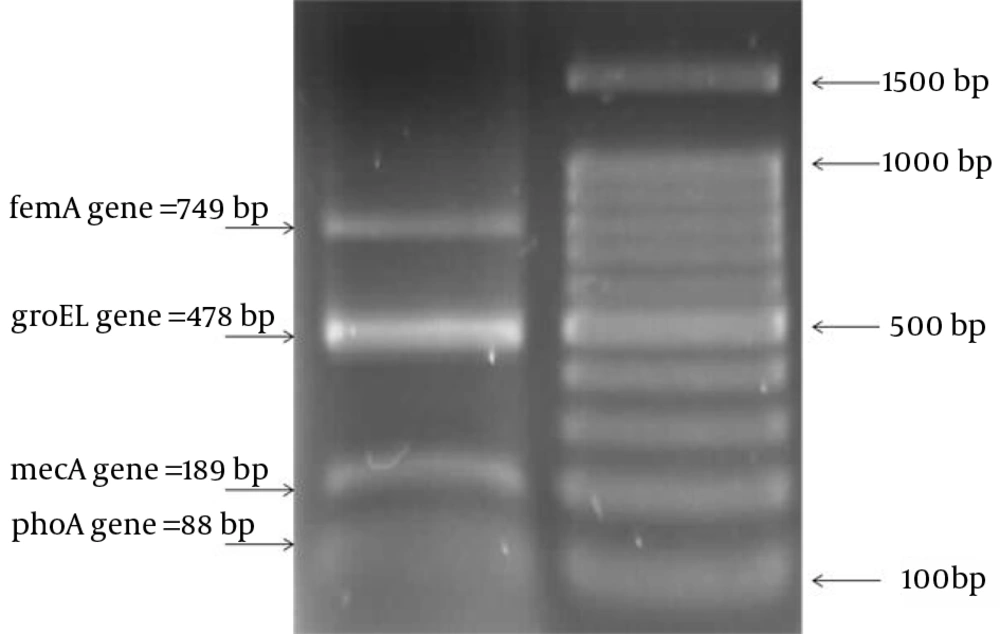
Identification of the 88 bp phoA gene among E. coli was 100%. Similarly, the 310 bp mecA fragment was obtained from all of the resistant S. aureus isolates after DNA amplification (Figure 1).
5. Discussion
Bacteremia is a serious condition leading to high expenses and a high level of admissions in hospitals. Moreover, bacteremia in cancer patients is associated with a high rate of morbidity and mortality; therefore, antibiotic treatment should be initiated immediately.
The results of our study from the blood cultures of cancer patients revealed that both Gram positive and negative bacteria were the causative agents of bacteremia and these bacteria were highly resistant to various clinical common antibiotics. In particular E. coli isolates were highly resistant to Penicillin, ampicillin, gentamicin, Cefoxitin, trimethoprim sulfamethoxazole and tetracycline, which is in consistent with other studies (10, 11); this is probably due to indiscriminate use of antibiotics and frequent use of antibiotics for both prophylactic and therapeutic treatment of hospitalized cancer patients.
Krcmery et al. (12) reported the etiology, risk factors, outcome and complications of bacteremia in 276 patients with solid tumors. They could isolate Klebsiella, Enterobacter, Pseudomonas aeruginosa, Acinetobacter spp. and Stenotrophomonas maltophilia. Finally, they concluded that mortality due to multiresistant Gram-negative bacteremias was higher in comparison to bacteremias due to susceptible organisms.
In our study, S. aureus isolates were highly resistant to gentamicin and sulfamethoxazole. Usually both these antibiotics are used for the treatment of serious S. aureus infections, particularly bacteremia (4, 13, 14). Using multiplex PCR, several studies (15, 16) reported the high prevalence of mecA gene in S. aureus strains; therefore, we suggest multiplex PCR as a rapid and reliable method, which gives reliable information to physicians for therapeutic management of bacteremia. Another interesting part of our study was detection of B. cereus from blood culture bottles, which is relatively uncommon causing blood stream infection in cancer patients. This could be confirmed by looking for groEL gene using multiplex PCR. Vigil et al. (17) reported the burden of E. coli causing bacteremia in cancer patients; furthermore, by Rep-PCR they confirmed the E. coli isolates as MDR.
Our results clearly indicate that all the bacterial isolates were associated with a high rate of resistance to various antibiotics and this resistant pattern could be confirmed by detection of a particular gene for each bacterium. Further studies should be performed to investigative the mechanism of resistance in bacterial isolates.