1. Background
Pseudomonas bacteria are Gram-negative, ambulant, aerobic, and viable bacteria, which normally form small colonization in the intestine and the skin flora. Pseudomonas aeruginosa is one of the important causes of both hospital and community acquired infections (1). The emergence of drug resistance in bacteria and its association with serious infectious diseases have increased at warning rates during the past several decades (2). Pseudomonas aeruginosa has remained the leading pathogen causing burn wound infection. It has been found as a major colonizer of burn wounds because it thrives on moist burn wound surfaces and survives well in hospital environments (3). The basic antibiotics with activity against P. aeruginosa strains are cephalosporins, carbapenems, monobactams, quinolones, aminoglycoside, and polymyxin. However, the emergence of drug resistant P. aeruginosa in burn wounds is becoming a challenging problem worldwide. The antibiotic resistance against these bacteria is categorized as multiple drug resistant (MDR) and pan-drug resistant (PDR) (4). Many studies have shown that MDR isolates exhibit resistance against three types of anti-pseudomonas drugs, and PDR show resistance against all types of antibiotics (5).
Several resistance mechanisms are found in bacterial pathogens. Drug efflux pumps play a key role in drug resistance and also serve other functions in bacteria. Active efflux is now recognized as an important component of bacterial resistance to most classes of antibiotics. This mechanism is mediated by efflux pumps, which are membrane-associated active transporters promoting the extrusion of toxic compounds, including antibiotics, from the cells (6, 7). Bacterial drug efflux pumps have been categorized to five families. Amongst these families, drug exporters belonging to the resistance-nodulation-division (RND) family play a key role in clinically relevant resistance in Gram-negative bacteria including P. aeruginosa. In total, six systems belonging to the RND family of transporters have been characterized in the pathogen, MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexXY, MexJK, and MexGHI-OprD (8). A MexAB-OprM pump, encoded by the mexAB-oprM gene, consists of two inner membrane-associated components, MexA and MexB, and an outer membrane component, OprM. It has been suggested that MexB and OprM function as an antibiotic-exporting component and an antibiotic exit outer membrane channel, respectively (9). With their partially overlapping substrate specificities, MexAB-OprM and MexXY play a key role in the natural resistance of P. aeruginosa to antibiotics. Overproduction of MexAB-OprM may lead to significant multidrug resistance in clinical isolates of P. aeruginosa (10).
2. Objectives
Due to the importance of drug resistance in P. aeruginosa strains, which is an increasing threat for patients with burn and wound infections, the present study was conducted with an aim to investigate the MDR and PDR rate in Pseudomonas aeruginosa in the region of the study, and detect efflux pump MexAB genes as a possible mechanism involved in resistance, by the PCR technique.
3. Materials and Methods
3.1. Bacterial Isolates and Strain Identification
In total 150 samples comprised of 75 samples of wound infections and 75 samples of burn infections were collected in a four-month period from April to September 2012 in Golestan and Taleghani burn hospitals, Ahvaz, Iran. The samples were obtained from equal numbers of male and female patients with an age range between 18 and 73 years.
The samples were cultured on appropriate culture media and grown colonies were identified as P. aeruginosa by applying standard biochemical tests, including cetrimide agar and Oxidization Fermentation (OF) media, reaction in triple sugar iron (TSI) agar, catalase test, indole production in SIM medium, and growth at 42°C (11). Pseudomonas aeruginosa ATCC 27853 was used as a standard strain for confirming the reactions.
3.2. Antimicrobial Susceptibility Testing
3.2.1. Disk Diffusion Method
The resistance pattern of the strains against the following antibiotics was determined by the Kirby Bauer disk diffusion method as per the Clinical and Laboratory Standard Institute (CLSI) guidelines (12): ofloxacin (5 mg), amikacin (30 mg), gentamicin (10 mg), ciprofloxacin (5 mg), piperacillin (100 mg), aztreonam (30 mg), polymyxin B (300 mg), carbenicillin (100 mg), ceftazidime (30 mg), imipenem (10 mg), meropenem (10 mg), ticarcillin Ticarcillin (75 mg , cefepime (30 mg), ceftriaxone (30 mg), cefotaxime (30 mg) and tetracycline (30 mg) [MAST Co., UK]. In brief, the standard bacterial suspension was prepared by growth of bacteria in Mueller Hinton Broth (Merck, Germany) with subsequent adjustment equal to 0.5 McFarland, and inoculated on Muller Hinton agar, and the antibiotic disks were placed on the plate at identical distances. Plates were incubated for 16 - 24 hours at 35°C. The diameter of the zone of growth inhibition was measured and compared to standard values.
3.2.2. Broth Microdilution Method
Broth microdilution test was used for determination of minimal inhibitory concentrations (MIC) of used antibiotics in accordance with CLSI guidelines. Briefly, the stock solution of antibiotic powders (Sigma Inc. Germany) was prepared in appropriate solvents, from which the necessary serial dilutions were made. The standard bacterial suspension equal to 0.5 McFarland was prepared and the antibiotic dilutions were added to bacterial suspensions in microtiter plate wells (SPL Life Co., Korea), and were incubated at 37°C for 24 hours. The MIC of different tested antibiotics was then determined according to standard CLSI breakpoints (12).
3.3. Polymerase Chain Reaction Amplification of the MexA and MexB Genes
DNA extraction was performed by the simple boiling method of bacterial colonies from all isolates that were identified as P. aeruginosa by phenotypic criteria (13).
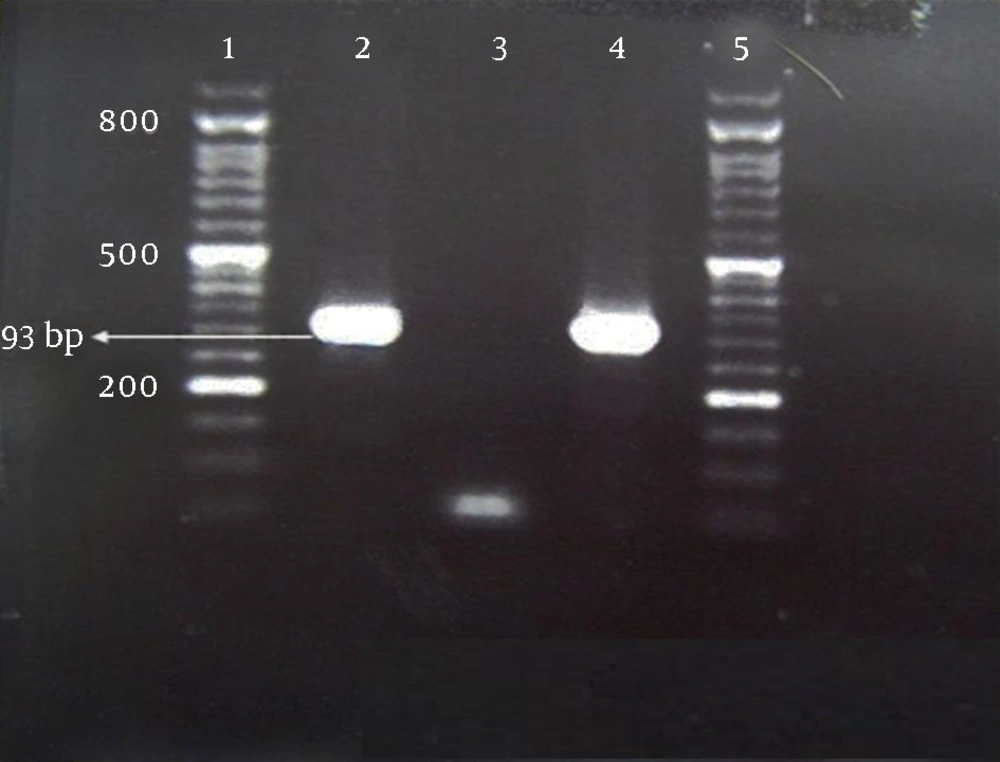
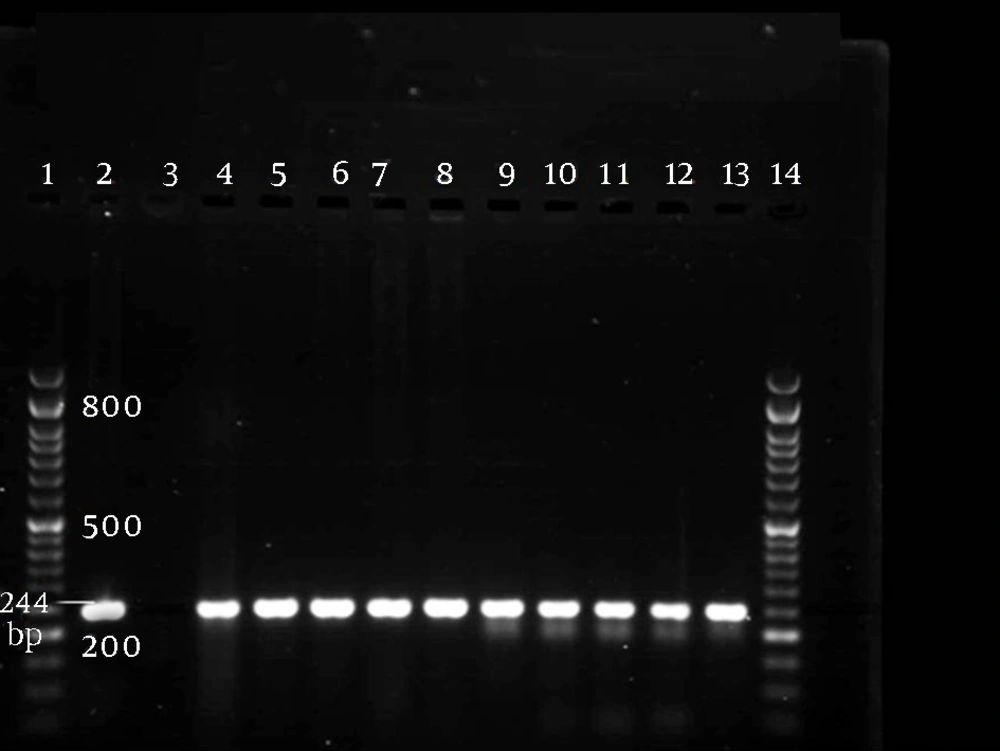
For PCR amplification, a set of primers for the MexA gene (5’- CGA CCA GGC CGT GAG CAA GCA GC -3’ and 5’- GGA GAC CTTCGCCGC GTT GTC GC -3’), and MexB gene (5’-GTGTTCGGCTCGCAGTACTC- 3’ and 5’- AACCGTCGGGATTGACCTTG- 3’), was used. These primers amplified a fragment of 293 base pairs (bp) and 244 bp of target genes, respectively (14).
DNA amplification was performed in a thermocycler nexus gradient (Eppendorf, Germany), in a final volume of 25 µL containing 10x PCR buffer, 5 mM MgCl2, 250 µM dNTPs, 0.4 µM of each primer, 0.125 Unit/µL Taq polymerase and 1 µL of template DNA (400 ng). All the reagents were purchased from Qiagen, Germany.
The PCR mixture was prepared in a final volume of 25 µL and consisted of 10X PCR buffer, 50 mM MgCl2, 10 mM dNTPs, 10 µM of each primer, 500 unit of Taq DNA polymerase, and 2 µL of template DNA. The amplification program consisted of initial denaturation at 94°C for five minutes, followed by 30 cycles of denaturation at 94°C for 40 seconds, annealing at 63.5°C for 30 seconds for the MexA gene, and 59°C for 30 seconds for the MexB gene, extension at 72°C for one minute and a final extension at 72°C for five minutes. Pseudomonas aeruginosa ATCC27853 was used as the positive control. The products were analyzed by agarose gel electrophoresis on 1.5% agarose (w/vol.) containing 0.5 mg/mL ethidium bromide (Qiagen, Germany). Results were recorded using the gel documentation system (Protein Simple, USA). A 100 bp DNA ladder was used as the size marker (Roche, Germany). The PCR products were sent for sequence analysis (Bioneer Co., Korea).
The SPSS software (SPSS Inc no. 13) was used for data analysis.
4. Results
According to the obtained results from qualitative susceptibility testing, P. aeruginosa strains showed more than 60% resistance to most of the tested antibiotics (Table 1). The most and least effective antibiotics against the bacteria were polymyxin B (96%) and cefotaxime (6%), respectively. According to the criteria determined for MDR (15), and PDR (4), 99 strains (66%) were identified as MDR, of which 53 (70.66%) belonged to burn and 46 (61.33%) originated from wound infections. There was only one strain isolated from a burn infection that was identified as PDR (0.67%), while none of the wound strains were PDR. The result of the microdilution test, as a quantitative method, confirmed the findings of the disk diffusion method. According to the results of this test, similar to the results of the disk diffusion method, polymyxin B was the most effective against P. aeruginosa isolates, with MIC of ≤ 2 µg/mL for sensitive strains (144 [96%]) and MIC of ≥ 8 µg/ml for resistant strains (6 [4%]). Besides, the highest resistance was seen against tetracycline, with no sensitive strain and MIC of ≥ 16 µg/mL for resistant strains (140 [93.93%]) as presented in Table 2.
| Antibiotic | Disk content, µg | Sensitive | Intermediate | Resistant |
|---|---|---|---|---|
| Ofloxacin | 5 | 38 (25.33) | 2 (1.34) | 110 (73.33) |
| Ciprofloxacin | 5 | 41 (27.33) | 3 (2) | 106 (70.67) |
| Gentamicin | 10 | 43 (28.67) | 4 (2.67) | 103 (68.66) |
| Imipenem | 10 | 53 (35.34) | 4 (2.67) | 93 (62) |
| Meropenem | 10 | 54 (36) | 3 (2) | 93 (62) |
| Ceftazidime | 30 | 48 (32) | 0 | 102 (68) |
| Amikacin | 30 | 60 (40) | 7 (4.67) | 83 (55.33) |
| Aztreonam | 30 | 30 (20) | 20 (13.33) | 100 (66.67) |
| Ceftriaxone | 30 | 30 (20) | 15 (10) | 105 (70) |
| Cefotaxime | 30 | 9 (6) | 32 (21.33) | 109 (72.67) |
| Cefepime | 30 | 30 (20) | 1 (0.67) | 119 (79.33) |
| Tetracycline | 30 | 140 (93.33) | 10 (6.67) | 0 |
| Ticarcillin | 75 | 44 (29.33) | 0 | 106 (70.67) |
| Piperacillin | 100 | 82 (54.67) | 0 | 68 (45.33) |
| Carbenicillin | 100 | 41 (27.33) | 2 (1.33) | 107 (71.34) |
| Polymyxin B | 300 | 144 (96) | 0 | 6 (4) |
aNote: the intermediate values are not shown in the table.
bData are presented as N. (%).
| Antibiotic | Antibiotics Serial Concentrations, µg/mL | MIC Resistance Interpretive Criterion, µg/mL | Resistant |
|---|---|---|---|
| Ofloxacin | 0.25 - 32 | ≤ 8 | 110 (73.33) |
| Ciprofloxacin | 0.25 - 32 | ≤ 4 | 106 (70.67) |
| Gentamicin | 0.5 - 64 | ≤ 16 | 103 (68.66) |
| Imipenem | 0.5 - 64 | ≤ 16 | 93 (62) |
| Meropenem | 1 - 128 | ≤ 16 | 93 (62) |
| Ceftazidime | 2 - 512 | ≤ 32 | 102 (68) |
| Amikacin | 1 - 128 | ≤ 64 | 83 (55.33) |
| Aztreonam | 1 - 128 | ≤ 32 | 100 (66.67) |
| Ceftriaxone | 2 - 512 | ≤ 64 | 105 (70) |
| Cefotaxime | 1 - 128 | ≤ 64 | 109 (72.67) |
| Cefepime | 1 - 128 | ≤ 32 | 119 (79.33) |
| Tetracycline | 1 - 128 | ≤ 16 | 0 (0) |
| Ticarcillin | 4 - 512 | ≤ 128 | 106 (70.67) |
| Piperacillin | 4 - 512 | ≤ 128 | 68 (45.33) |
| Carbenicillin | 4 - 512 | ≤ 512 | 107 (71.34) |
| Polymyxin B | 0.25 - 32 | ≤ 8 | 6 (4) |
aData are presented as No. (%).
According to the PCR technique, all MDR and PDR strains identified by traditional susceptibility methods (66.6%), contained MexA (Figure 1) and MexB (Figure 2) genes. We noticed a high resistance rate of isolates to fluoroquinolones (> 70%), carbapenems (> 60%), aztreonam (> 66%) and ticarcillin (> 70%), with a possible role of these genes in the isolates resistance capacity.
5. Discussion
Antibacterial resistance continues to be a global public health concern, and threatens the effectiveness of antibacterial therapy and challenges the efforts for developing novel antibacterial agents. Drug efflux pumps play a key role in drug resistance and also serve other functions in bacteria including P. aeruginosa (6, 16).
In this study we observed high levels of resistance to all antimicrobial agents commercially available in Iran, along with a rate of 66% MDR amongst P. aeruginosa isolates obtained from two teaching hospitals of Ahvaz, Iran. Although this rate was higher in strains isolated from burn samples (70.66%), compared to wound samples (61.33%), the difference was not statistically significant. Additionally, as shown in Table 1, more than 70% of the poly resistance was seen against seven antibiotics and resistance to 12 antibiotics was more than 60%. In a similar study, Pellegrino et al. reported 32% and 40% co-resistance to 8, 9, or 10 antimicrobial agents, at two different centers (17). Since their report dates back to about ten years ago, the lower rate of detected resistance compared to our finding may be expected. As a matter of fact, antibiotic resistance has increased dramatically around the world in the recent years. Higher antibiotic resistance compared to the present study was reported by Delpano et al. Dappano et al. In their study 100% resistance to cefepime, meropenem, imipenem, gentamicin, tobramycin, and ciprofloxacin, and 83% - 94% to other tested antibiotics were reported by using MIC test (18).
The frequency of P. aeruginosa resistance to ciprofloxacin, amikacin and gentamicin isolated from burn centers of Tehran, Iran, was over 85% in the study of Rastegar Lari et al. (19), which was higher compared to our findings. The main reason for this disagreement is that the samples of our study were not entirely collected from burn wounds, since the bacteria recovered from burn wounds harbor more antimicrobial resistance, suggesting that anatomic habitat selection was associated with adaptive resistance to antimicrobial drugs.
In concordance with this work, in a recent study from Iran, a rate of 60% for MDR in P. aeruginosa isolates from an intensive care unit was reported (20).
In MDR bacteria, the over-expression of efflux pumps that expel structurally-unrelated drugs, contributes to the reduced susceptibility, by decreasing the intracellular concentration of antibiotics (21). Active efflux is now recognized as an important component of bacterial resistance to most of classes of antibiotics. This mechanism is mediated by efflux pumps, which are membrane-associated active transporters promoting the extrusion of toxic compounds, including antibiotics, from cells (7).
Several studies have focused on the susceptibility patterns of P. aeruginosa isolates to carbapenems, which still are among drugs of choice for treatment of related P. aeruginosa infections.
Since resistance to many important clinical in-use antibiotics, including B-lactam antibiotics and fluoroquinolones, is mediated by the MexAB-OprM efflux pump (8), we chose this pump to investigate the existence of encoding genes. The present study showed the presence of genes encoding the MexAB-OprM efflux pump, and more than 65% resistance of all tested isolates to carbapenems, ticarcillin, aztreonam and fluoroquinolones (ofloxacin and ciprofloxacin), which indicates of the possible role of this efflux pump in such high resistance. We previously reported a rate of 38% MDR with lower resistance of 41% to carbapenems (21). Pseudomonas aeruginosa can become resistant to carbapenems by both intrinsic (mutation-driven) and transferable (β-lactamase-based) mechanisms. It is important to mention that several unrelated antibiotic molecules, e.g. imipenem and fluoroquinolones, are able to select for the over-expression of efflux pumps in originally susceptible patient isolates (22).
In the study of Fuste et al. the mechanisms leading to carbapenem resistance of an MDR P. aeruginosa clone were investigated. Their results showed an overexpression of the MexAB-OprM efflux pump, and a functional MexXY-OprM was detected in all isolates (23).
In a similar study by Yi et al. 27 out of 49 P. aeruginosa strains which were resistant to carbapenem (86.6% to imipenem and 44.4% to meropenem), over-expressed the mexAB-OprM (24). In a study conducted by Lee and Ko, among 57 carbapenem-resistant P. aeruginosa isolates, 41 (71.9%) showed MexAB-OprM and AmpC over-expression (56.1% and 47.4%, respectively) (25). Overall, studies that represent drug resistance in strains with over-expression of efflux pumps, especially in isolates from burn patients and patients with decreased immune defense, have increased during the past years.
In the area of the present investigation, resistance to carbapenems has raised from 41% in 2008 to 62% in 2012. Additionally, the MDR rate has increased from 38% to 62%, which shows the dramatic ineffectiveness of antimicrobial agents in the treatment of P. aeruginosa isolates. Fortunately the rate of PDR was still low in hospitals under investigation of the present study, thus a strategy should be made to limit the MDR rate and prevent the PDR strains to be raised especially in burn units. In this study we only investigated the presence of one of the numerous genes, which encode efflux pumps. We can not confirm for sure that the presence of the genes reveal the existence a functional MexAB efflux pump and mediate resistance, since confirmation needs much more experimental work. However, this work is a preliminary point to investigate the genetic basis of efflux pumps-mediated resistance in P. aeruginosa, which should be continued in the future.
This study represented an increasing rate of MDR P. aeruginosa in burn and wound samples. Efflux MexAB genes were detected in all MDR and PDR strains. The P. aeruginosa strains isolated from burn cases showed higher drug resistance, while PDR resistance was only noted in a burn samples.
In conclusion, the present study demonstrated the relatively high MDR P. aeruginosa in burn and wound samples obtained from two university hospitals. The P. aeruginosa strains isolated from burn cases showed higher drug resistance while PDR resistance was only noted in a burn sample. Efflux MexAB genes were detected in all MDR and PDR strains. By understanding the exact mechanisms involved in drug resistance, the management and strategy of infection therapy could be improved in regions with known higher antimicrobial resistance.