1. Background
The transcriptome analysis of cells and tissues is the best method for determining their functions (1). The venom of scorpions is composed of peptides that are useful in biological and medical research (2, 3). Because some venom peptides affect a wide variety of physiological processes, including cell excitability, the regulation of the heartbeat, muscle contraction, neurotransmitter release, hormonal secretion, signal transduction, and cell proliferation, they have the potential to be used in drug development (4). The molecular identification of venom peptides could provide a framework for pharmaceutical, medical, and biological studies. One of the best ways to identify information about these peptides is to access the genomic sequences coded in these peptides and then clone them to a suitable host so they can be expressed in a large quantity. cDNA library construction is the best method for transcriptome analysis and cloning or expressing the venom peptide sequences. In cDNA library construction, the total RNA of a tissue converts to ds cDNA and then inserts into a desired vector. Vectors with ds cDNA then transform into a suitable competent host cell. After, the cells grow in agar media. Finally, in isolating every clone in the library, the primary aim outlined above is obtained (5-8).
Using bacterial hosts for cDNA library construction and for the cloning of genes has been prevalent in the past (9). Because of its ease of use, benign characteristics, and high adaptation to various conditions, Escherichia coli (E. coli) is one of the best bacterial hosts for cloning (10).
In this study, we set up a cloning system using the DH5α strain of E. coli as a host for transcriptome analysis and the construction of cDNA libraries from the venom glands of two Iranian scorpions, Odonthubuthus doriae (O. doriae) and Mesobuthus eupeus (M. eupeus). These genera of the Buthidae family are two of the most dangerous species of scorpion. O. doriae is mostly found in the central area of Iran, while M. eupeus is found in almost all areas, as it is the most abundant scorpion in Iran.
2. Objectives
The transcriptome analysis and cDNA library construction from the venom glands of these Iranian species and other Iranian scorpions have not been performed until now. We set up and improved an existing kit to make the transcriptome analysis and cDNA library construction of the venom glands of any other scorpion species possible.
3. Methods
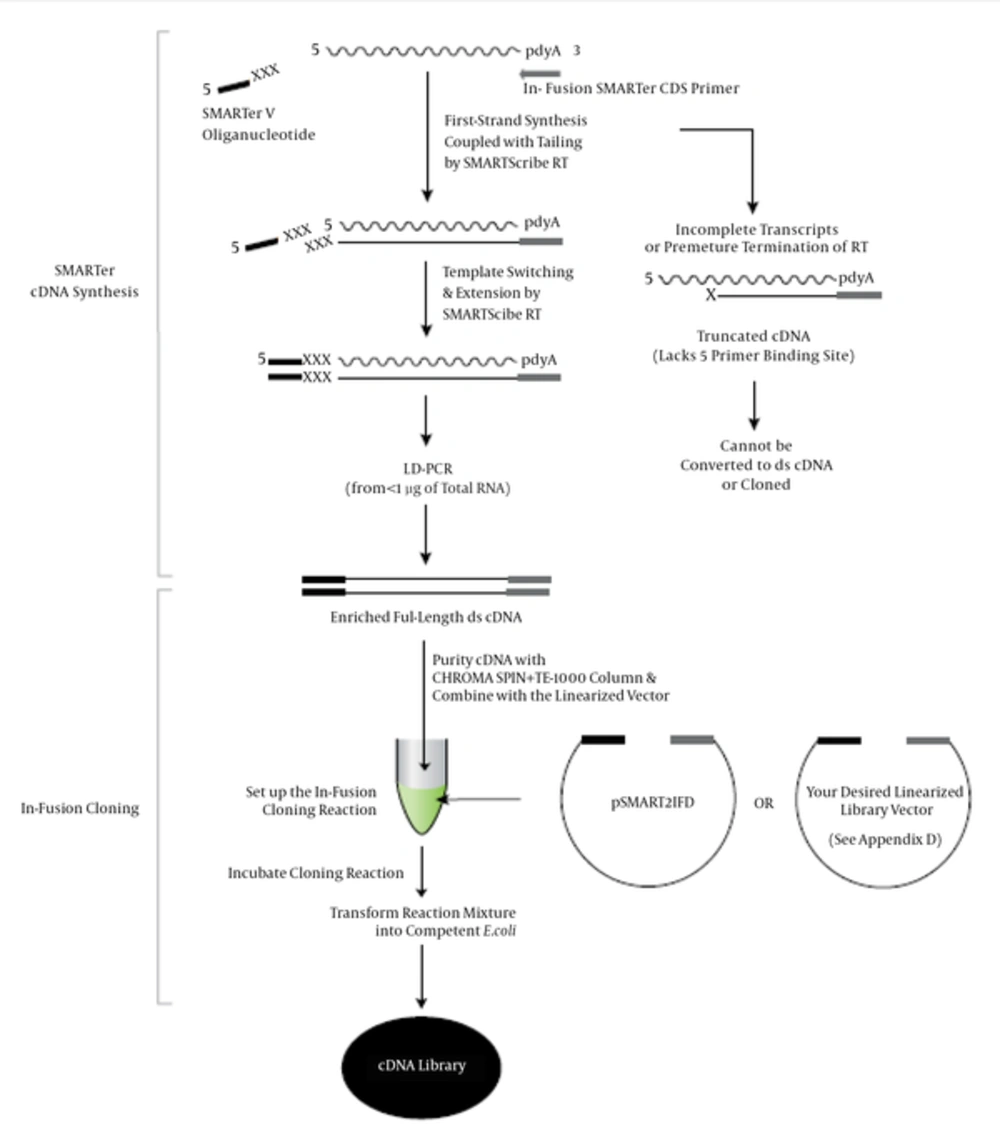
To construct the cDNA libraries, we used and improved the In-Fusion® SMARTer™ Directional cDNA library construction kit from Clontech laboratories (Figure 1). The kit provides a dependable method for producing high-quality, full-length cDNA libraries. It utilizes two of Clontech’s most innovative technologies: SMART™ cDNA Synthesis and In-Fusion® HD Cloning. SMARTer™ technology enables the full-length cDNA synthesis of poly A+ or total RNA in nanograms, and In-Fusion® Cloning makes the cloning of SMARTer™ cDNA libraries possible in any location within any vector, including the pSMART2IFD Linearized Vector provided by the kit. Isolated clones from SMARTer™ libraries can be transferred directly to any linearized expression vector without needing compatible restriction sites for functional analysis.
As shown in Figure 1, this kit uses electrocompetent cells for transformation. However, we optimized the kit using chemically competent cells in this project.
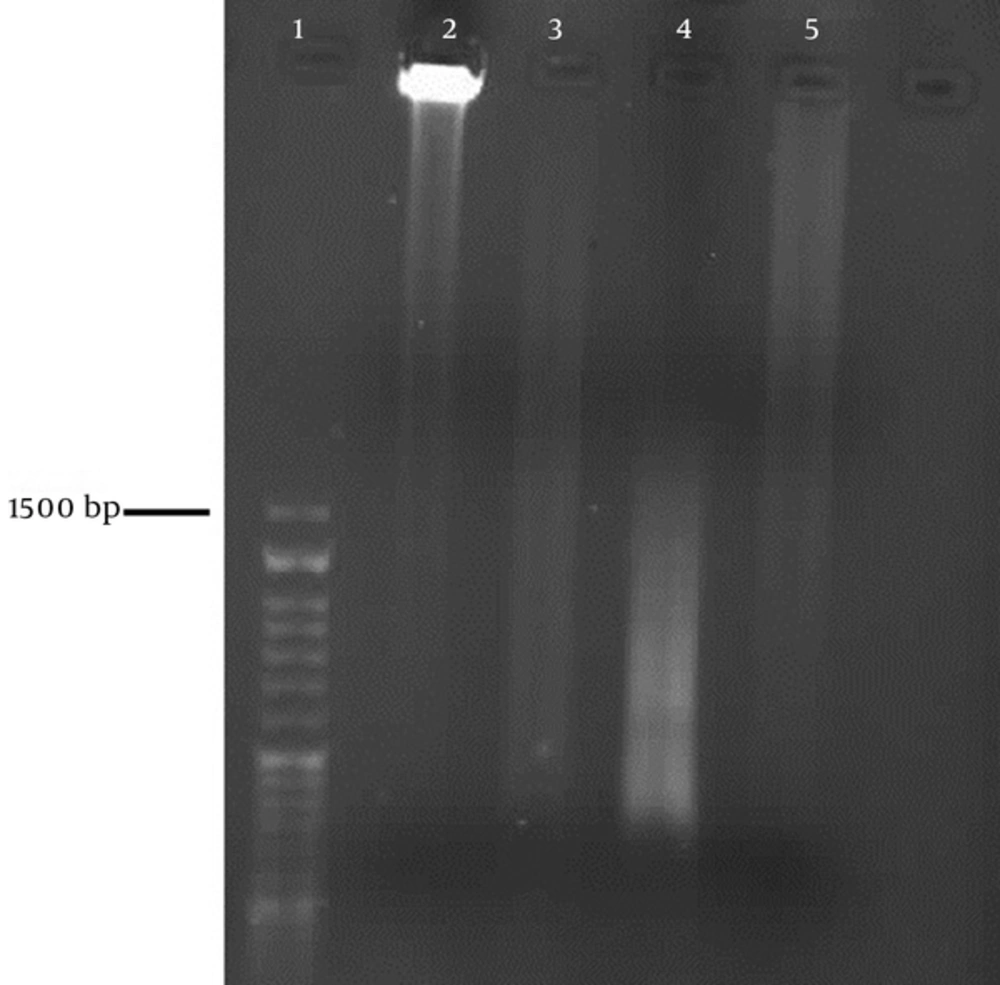
First, numerous O. doriea and M. eupeus scorpions were obtained from the central and south areas of Iran, respectively. The optimum number of each species was 6 - 10. Their telsons were cut 3 days after milking them. Total RNA samples were extracted from mashed and homogenized telsons using the RNeasy® plus mini kit (Qiagen). After checking the concentration of the isolated RNA samples using a nanodrop, cDNA was synthesized by the PCR program used in the In-Fusion® SMARTer™ Directional cDNA library construction kit. In this step, the specific adaptors ligated, and cDNA was synthesized from mRNA. The synthesized cDNA samples were purified by the CHROMA SPIN columns included in the kit, and the quantity was obtained using a nanodrop. The quality of the synthesized cDNA was checked by running 3 µL of the cDNA solution mixed with a 1.2% agarose/EtBr gel in a 1X TAE buffer. Typical ds cDNA synthesis results using PCR should have a moderately strong smear of cDNA (from 0.5 - 3 kb).
The cDNA samples were inserted into the pSMART2IFD Linearized Vector provided by the kit. In ligation reactions, maximum cloning efficiency is achieved when using a 2: 1 molar ratio of insert to vector. In this project, the optimum value was achieved at 2.5 µl (60 - 70 ngr/µL) from the cDNA insert to 0.5 µL (150 ngr/µL) of the vector. The cloned vectors transformed into DH5α strain host cells of E. coli.
Before the transformation, competent cells were prepared by heat shock treatment and cold Calcium Chloride (CaCl2) instead of using electrocompetent cells, as recommended by the kit. The transformed bacteria were revived in S.O.C. culture media on a shaking incubator at 37°C for 1 hour.
3.1. Preparing Chemically Competent Cells
To prepare the chemically competent cells, a single colony was inoculated into 10 mL of LB broth culture media in a 50 mL falcon tube at 37°C. The next morning, 1 mL of overnight culture was inoculated in 100 mL of LB mixed with 250 mL of Erlenmeyer. It was shaken at 37°C until the OD600 reached 0.45 - 0.65.
Then, 5 mL of pure culture was placed on a 15 mL falcon and set on ice for 10 minutes. The bacterial cells were pelleted at 6000 rpm for 5 minutes, and all the liquid above the pellet was carefully removed. The pellet was resuspended in 300 μL of cold CaCl2 (0.1 M), and then the tube was placed on ice for 30 minutes.
After, the tubes were centrifuged at 6000 rpm for 5 minutes. All the liquid above the pellet was removed, and the pellet was resuspended in 200 μL of CaCl2. The tubes were placed on ice for 30 minutes, and then the tubes were centrifuged at 6000 rpm for 5 minutes. All the liquid above the pellet was removed, and the pellet was resuspended in 100 μL of CaCl2. Next, the tube was placed on ice for 7 minutes.
3.2. Bacterial Transformation Protocol
To transform the bacteria, we added 100 μL of competent cells to 10 μL of the plasmid created in the cloning step and incubated the mixture on ice for 30 min. Then, a heat shock was performed at 42°C for 90 seconds. The mixture was incubated on ice for 2 minutes. After, we added 1 mL of liquid S.O.C. media and incubated the mixture at 37°C for 1 hour in a shaking incubator.
Next, we spread 50 μL of the mix onto a 90 mm LB agar plate containing 100 μg/mL of Ampicillin, 1 mM of IPTG, and 75 μg/mL of X-Gal. We incubated the plates at 37°C overnight. At this stage, the cDNA libraries were constructed.
To screen the cDNA libraries, blue-white screening was first used, and cDNA was inserted into the white colonies. To determine the percentage of recombinant clones, a colony PCR method was designed with specific primers for two sides of the insertion site into the vector provided by the In-Fusion® SMARTer™ Directional cDNA library construction kit. TCACACAGGAAACAGCTATGA was used as the forward screening primer, and CCTCTTCGCTATTACGCCAGC was used as the reverse screening primer.
The PCR programs include one with 3 cycles (at 95°C for 30 seconds, 65°C for 30 seconds, or 68°C for 3 minutes) and one with 25 cycles (at 95°C for 30 seconds or 68°C for 3 minutes). For the PCR, 3 μL of PCR product was run in a 1.2% TAE/agarose gel with DNA size markers. Typically, bands > 700 bp in size are used for colonies containing inserts.
To confirm the construction of the scorpion cDNA libraries, every white colony was selected and cultured separately as one transcript in 10 mL of LB broth media containing 100 μg/mL. After an overnight culture, the plasmid DNA was isolated from the liquid culture using the QIAprep® Spin Miniprep Kit (Qiagen). Then, the inserted cDNA was sequenced by specific primers designed for the two sides of multiple cloning sites into the vector. Finally, every obtained sequence was compared with those in the GeneBank database using the BLAST software from the NCBI website (http://www.ncbi.nlm.nih.gov).
4. Results
The optimum amount of ds cDNA in this project was obtained at 60 - 70 ngr/µL. The smears of ds cDNA samples synthesized by gel electrophoresis for both the scorpions are depicted in Figure 2. The smears ranged from 0.5 to much more than 1.5 kb.
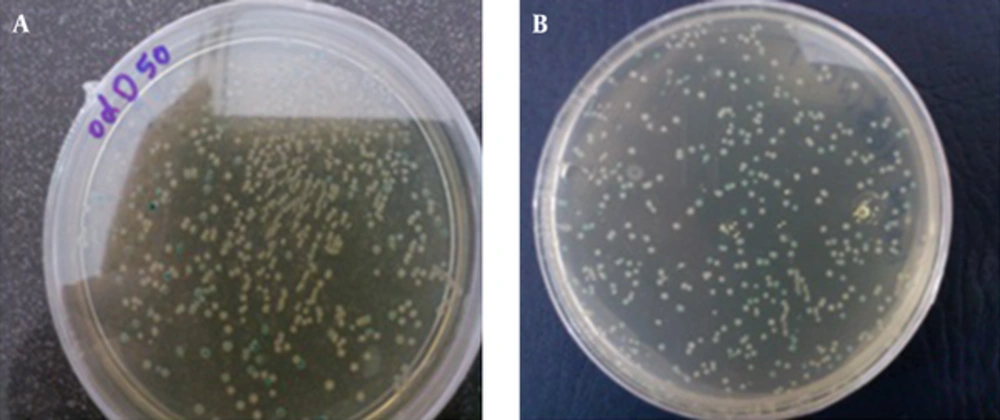
Over 300 blue and white colonies of bacteria were observed on the plate 12 - 18 hours after the transformation. Most of the colonies were white; therefore, a large number of bacterial cells transformed with inserts. Every clone had one cDNA exponential to its transcript.
To obtain the best yield, it is important that the cells used for the transformation are in the early log phase of growth. We obtained the best result when the OD600 range reached 0.45 - 0.65. More than 300 blue and white colonies had vectors on the plate containing the transformed cells cultured overnight (Figure 3). The white colonies of bacteria with inserts had vectors; however, the blue colonies without inserted cDNA had vectors.
Screening the white colonies and the specific primers designed for both sides of the insertion site using PCR showed 61 unique sequences in the cDNA library of M. eupeus and 50 unique sequences in the cDNA library of O. doriae. In the bioinformatics analysis of the white colonies, the obtained cDNA sequence showed similarities to scorpion toxins, enzymes, and other scorpion peptides, confirming the construction of the cDNA libraries of the two Iranian scorpions’ venom glands.
5. Discussion
Because cDNA are different sizes and have different coded protein components of venom, the results of ds cDNA synthesis using PCR with agarose gel should have smears. This is consistent with the picture obtained from our cDNA electrophoresis.
Many factors affect cDNA transformation, cloning, and library construction; in this study, we changed and optimized some of these factors to obtain the best possible results. The bacterial culture medium used is an important factor that affects the transformation efficiency. Fierro et al. (11) and Maeda et al. (12) suggested that S.O.C media is richer than Lb media in the growth of bacteria for the preparation of competent cells, as the bacteria can grow faster. Therefore, we used S.O.C culture media to culture the transformed bacteria in the resuscitation step before culturing it using LB agar.
Tu et al. (13) proposed that different strains of E.coli have different growth characteristics; therefore, the optimal OD600 range varies for the preparation of competent cells. In their study, the OD600 range used for the DH5α strain was reported at 0.145 - 0.45. However, in the current study, the optimum OD600 range for the preparation of competent cells from the DH5α strain was found to be 0.45 - 0.65, which is higher than the value obtained before.
The CaCl2 heat shock method was first introduced by Mendel et al. (14). It was further developed by Inoue et al. (9) and Panjaet al. (15) and improved by other researchers. In the current study, different rotation times and speeds were used after each step of incubation than what was previously reported. Under these conditions, we obtained the highest transformation efficiency.
Electrocompetent cells were produced during the electroporation process using Microelectromechanical systems. Although this method has a high efficiency, the device and the raw materials used in this process are costly. The preparation of chemically competent cells is time-consuming and may require a detailed set up; however, it is less costly and, in some cases, more practical. Therefore, chemically competent cells were prepared in this study, and an acceptable yield was obtained. Using the settings described in this study, the cDNA library of any scorpion can be synthesized with a low cost and less time than ever before.
The cloning reaction is outlined by the manual of the kit. Based on the manual, the required vector and cDNA amounts are based on the volume. However, another part of the manual mentions that maximum cloning efficiency is achieved when using a 2: 1 molar ratio of insert to vector. Similarly, in this study, the best results were obtained when the cDNA concentration was twice the vector concentration in the cloning reaction.
Singh et al. pointed out that the transformation of bacterial cells exposed to CaCl2 occurs before heat shock treatment but that its efficiency is higher after heat shock treatment. When the bacterial cells were incubated on ice following heat shock treatment, the transformation efficiency increased 24-fold (16). In our research, we used both CaCl2 and heat shock treatment to obtain acceptable results.
The colonies obtained in the current cDNA libraries were mostly white. Therefore, the technique and set up used were very efficient. Ultimately, there are 61 raw nucleotide sequences of M. eupeus venom and 50 raw nucleotide sequences of O. dorieae in our constructed cDNA libraries, as a result of new putative peptides. Similarity, the insert sequences of the white colonies with venom components assure us of the correctness of our cDNA libraries. According to the potential use of scorpion venom for new drug development (2), the obtained cDNA libraries can be a platform for studying protein expression, crystallography, nuclear magnetic resonance, and pharmacological effects on laboratory animals.