1. Background
Urinary tract infection (UTI) is one of the most common infections among outpatients visit medical centers (1). Escherichia coli is the most common etiologic agent of UTI and has been isolated in 75% to 90% of cases (2). The incidence of UTIs caused by strains of community acquired extended spectrum β-lactamase-producing (ESBL) E. coli is globally increasing (3, 4). In recent years, fluoroquinolones have been used as alternatives for empirical treatment of UTIs caused by E. coli (5). A systematic review in Iran revealed that 28.2% of E. coli isolates from UTI were resistant to ciprofloxacin (6). The quinolones inhibit bacterial DNA synthesis by binding to the topoisomerase II (DNA gyrase) and topoisomerase IV enzymes (7). The topoisomerase II consists subunits of A and B encoded by gyrA and gyrB, respectively. Similarly, the topoisomerase IV also consists of C and E subunits encoded by parC and parE, respectively (7). DNA gyrase in E. coli is the main target of quinolones (8), an enzyme which is essential for preserving the bacterial DNA topology. The second target of quinolones in gram-negative bacteria is the topoisomerase IV involves the separation of daughter cells’chromosomes at the end of DNA replication cycles (8, 9). The widespread use of fluoroquinolones in empiric treatment of UTI has resulted in resistance among E. coli strains (5). So far, 3 basic mechanisms of resistance to fluoroquinolones have been identified: 1, mutations in quinolone resistance determining region (QRDR) of topoisomerase genes; 2, decreasing the concentration of antibiotics within bacterial cells by efflux pumps or decreased expression of the porins; and 3, acquisition of plasmid- mediated resistant genes (10, 11). The most important mechanisms of resistance to quinolones in E. coli is mutations in DNA gyrase and topoisomerase IV (12). Mutations usually take place within the N-terminal regions of gyrA and parC subunits, which are more common than mutations in gyrB and parE (7). In E. coli, gyrA mutations usually occur at serine-83, which is substituted by leucine or tryptophan and causes high resistance to quinolones. However, the replacement of serine by alanine causes a lower resistance level (8, 13). Another common mutation is the substitution of aspartate-87 by asparagine and/or valine (13). Mutations in parC usually occur at 78, 80 and 84 positions, with the replacement of glycine by aspartate, serine by arginine, or isoleucine and glycine by aspartate, respectively (14). The amino acid substitutions can have a significant effect on the minimal inhibitory concentration (MIC) of fluoroquinolones. Research has shown that 3 or 4 mutations are required for high level of resistance (15, 16). The present study aimed at identifying mutations in gyrA and parC and the correlation between fluoroquinolone MIC and the number of mutations among ESBL producing E. coli.
2. Methods
2.1. Bacterial Isolates
In this cross-sectional study, 240 isolates of E. coli were isolated from urine of patients with urinary tract infections during 2014 and 2015 in Kermanshah, west of Iran. Urinary tract infection was defined as 105 or more bacteria per 1 mL of urine in people clinically suspected of having urinary infection (17). Isolates were identified using bacteriological and biochemical tests (18).
2.2. ESBL Screening of Isolates
For screening the ESBL production of isolates, the combined disc test was used according to the Clinical and Laboratory Standards Institute (CLSI) protocols (18). Briefly, the combined discs of ceftazidime (30 µg) plus clavulanic acid (10 µg) and cefotaxime (30 µg) plus clavulanic acid (10 µg) (MAST, England) were used in a standard disk diffusion assay. If the diameter of inhibited growth zone around the combined discs for an isolate was at least 5 mm or more than the zone for single disc of the same antibiotic, then, the isolate was considered ESBL-producer (19). The strain of ESBL-producing E. coli ATCC 35218 was used for quality control.
2.3. Antibiotic Susceptibility Testing
The susceptibility of E. coli isolates to ciprofloxacin and levofloxacin (MAST, England) was first assessed by the standard disk diffusion method according to the CLSI guidelines (19). Then, the MIC for ciprofloxacin and levofloxacin (Sigma, USA) was determined by the broth microdilution method using CLSI criteria (19). Briefly, a serial dilution of ciprofloxacin and levofloxacin was prepared in Mueller Hinton broth containing 5 × 106 CFU/mL bacteria. The culture tubes were incubated at 37°C for 18 hours and the lowest concentration of antibiotic with no visible bacterial growth was defined as the MIC. E. coli ATCC 25922 strain was used as the quality control. The range of antibiotic concentrations was from 0.015 to 1024 µg/mL. The CLSI breakpoints were used for ciprofloxacin (susceptible ≤ 1 µg/mL; resistant ≥ 4 µg/mL) and levofloxacin (susceptible ≤ 2 µg/mL; resistant ≥ 8 µg/mL).
2.4. DNA Amplification and Sequencing
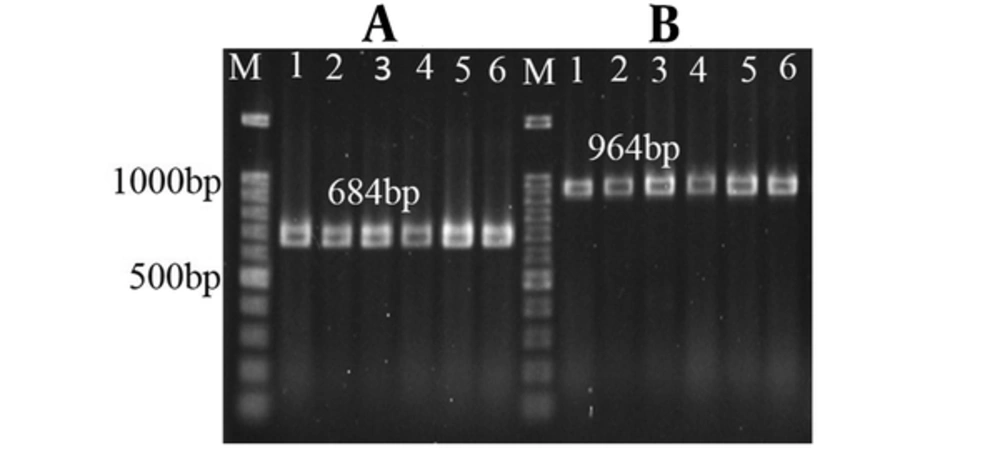
Bacterial DNA was extracted using genomic DNA purification kit (SinaClon, Iran). The QRDR of parC and gyrA genes in 45 isolates resistant to both antibiotics (ciprofloxacin and levofloxacin) was amplified by PCR (Table 1). After electrophoresis of PCR products on 1% agarose gel and staining with ethidium bromide, the DNA bands were visualized by GelDoc apparatus (BioRad, USA).
All PCR products for parC and gyrA genes were purified by kit and sequenced (SinaColon, Iran). The sequence was performed using an ABI 3730XL DNA analyzer apparatus (Macrogen Inc., Korea). Sequence data were analyzed for homology with genetic data using the national center for biotechnology information GenBank database (http:/www.ncbi.nlm.nih.gov/BLAST/).
2.5. Statistical Analysis
All data were analyzed using SPSS Version 20 for the correlation between gene mutations and ciprofloxacin and levofloxacin resistance by chi square or Fisher’s exact test. Statistical significance was defined as P value less than 0.05.
3. Results
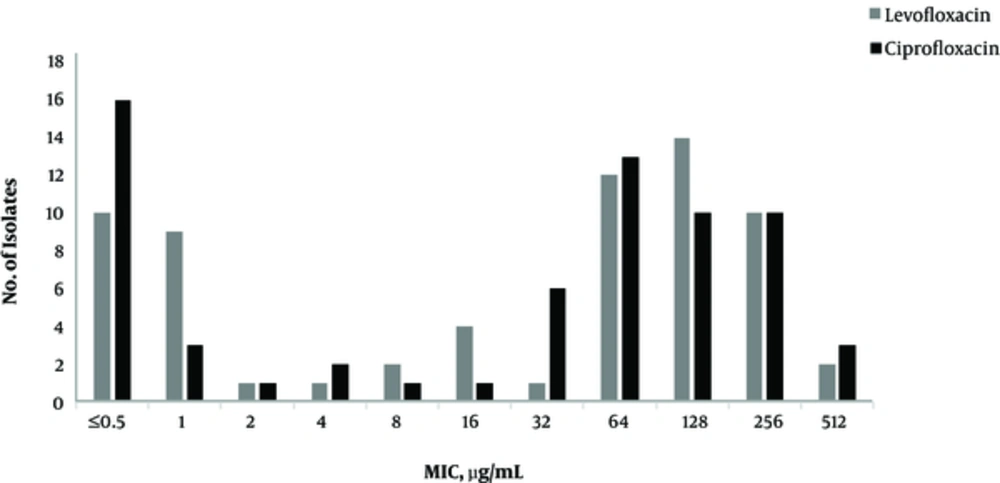
Of 240 isolates, 66 (27.5%) were ESBL positive. The isolates belonged to 57 women (86.4%) and 9 (13.6%) males with the average age of 43.5 ± 22 years. In terms of age distribution of the patients, 12 (18.2%), 14 (21.2%) and 40 (60.6%) were under 20, 21 to 40 and above 40 years, respectively. The antibiotic susceptibility and MIC of ciprofloxacin and levofloxacin for 66 ESBL producing E. coli isolates are presented in Table 2 and Figure 1. Of these 66 isolates, 45 (68.1%) were resistant to both ciprofloxacin and levofloxacin. The amplification results of gyrA and parC genes are displayed in Figure 2.
| Fluoroquinolones | Antibiotic Susceptibility of Isolates | ||||
|---|---|---|---|---|---|
| Susceptible | Intermediate | Resistance | MIC50 | MIC90 | |
| Ciprofloxacin | 20 (30.3) | 1 (1.5) | 45 (68.1) | 64 | 256 |
| Levofloxacin | 20 (30.3) | 1 (1.5) | 45 (68.1) | 64 | 256 |
aValues are expressed as No. (%).
DNA sequence analysis of the QRDR of gyrA and parC showed mutations in gyrA and parC in 45 isolates resistant to both antibiotics. All isolates resistant to fluoroqinolones showed mutations in gyrA at codon 83 (C → T Transition of codon TCG) and resulted in substitution of serine by leucine. A total of 42 (93.3%) isolates showed mutations at codon 87 (G → A Transition of codon GAC) and resulted in the substitution of aspartic acid by asparagine.
Mutations at 80 and 84 amino acid positions were found in QRDR of parC. In 42 (93.3%) isolates, a mutation was found at codon 80, in 40 cases serine was substituted by isoleucine (G → T Transversion of codon AGC), and in 2 cases serine was substituted by Leusine (AGC → CTT). A mutation at codon 84 of parC was found in 26 (57.7%) isolates, in 25 cases of them the A → T transposition of codon GAA was detected that resulted in substitutions of glutamic acid by valine, and in 1 case the G → A transposition of codon GAA resulted in the substitutions of glutamic acid by lysine. A mutation outside QRDR of parC was also detected in 22 (48.8%) isolates where C → T transposition in GCA codon at 192 position resulted in the substitution of alanine by valine amino acid. The nucleotide sequence data of ParC have been deposited into the GenBank under the accession number of Ky308189.
The correlation of mutations in gyrA and parC with the MIC of isolates is demonstrated in Tables 3 and 4. Isolates with a single mutation in gyrA and no mutations in parC showed the lower MIC breakpoints compared to isolates with mutations in QRDR of both parC and gyrA genes, which were statistically significant for both ciprofloxacin (P = 0.008) and levofloxacin (P = 0.026).
| Mutations in the QRDR | No. of Isolates | No. of Isolates with MIC (≥ 4 µg/mL) to Ciprofloxacin | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | ||||||||||||
| Ser83 | Asp87 | Ser80 | Glu84 | Ala192 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | |
| Leu | Asn | Ile | Val | - | 3 | 1 | 1 | 1 | |||||
| Leu | Asn | Ile | - | - | 15 | 1 | 1 | 7 | 2 | 4 | |||
| Leu | - | - | - | - | 3 | 2 | 1 | ||||||
| Leu | Asn | Ile | Val | Val | 20 | 3 | 6 | 5 | 4 | 2 | |||
| Leu | Asn | Ile | - | Val | 1 | 1 | |||||||
| Leu | Asn | Ile | Lys | - | 1 | 1 | |||||||
| Leu | Asn | Leu | Val | - | 1 | 1 | |||||||
| Leu | Asn | Leu | Val | Val | 1 | 1 | |||||||
Abbreviations: Ala, Alanine; Asn, Asparagine; Asp, Aspartic Acid; CLSI Breakpoint for Ciprofloxacin Resistant; Glu, Glutamic Acid; Ile, Isoleucine; Leu, Leucine; Lys, Lysine; Ser, Serine; Val, Valin.
| Mutations in the QRDR | No. of Isolates | No. of Isolates with MIC (≥ 8 µg/mL) to Levofloxacin | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | |||||||||||
| Ser83 | Asp87 | Ser80 | Glu84 | Ala192 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | |
| Leu | Asn | Ile | Val | - | 3 | 1 | 2 | |||||
| Leu | Asn | Ile | - | - | 15 | 1 | 1 | 4 | 4 | 5 | ||
| Leu | - | - | - | - | 3 | 2 | 1 | |||||
| Leu | Asn | Ile | Val | Val | 20 | 1 | 7 | 8 | 2 | 2 | ||
| Leu | Asn | Ile | - | Val | 1 | 1 | ||||||
| Leu | Asn | Ile | Lys | - | 1 | 1 | ||||||
| Leu | Asn | Leu | Val | - | 1 | 1 | ||||||
| Leu | Asn | Leu | Val | Val | 1 | 1 | ||||||
Abbreviation: CLSI, Breakpoint for Levofloxacin Resistance.
4. Discussion
Following the extensive use of fluoroquinolone in the past few decades, resistance to this group of antibiotic has increased in E. coli. Given the cotransmission of resistance genes, research showed that the ESBL positive isolates are more resistant to fluoroquinolones than non-ESBL isolates (22, 23). Our results revealed a high resistance rate to ciprofloxacin and/or levofloxacin among ESBL positive isolates of E. coli, which is consistent with the results reported for fluoroquinolones by a number of studies in Iran (64.2%), Canada (79.4%), Egypt (60%) and Syria (65.8%) (24-27). Given the fact that only ESBL positive isolates were used in our study, it is reasonable to expect a higher antibiotic resistance compared to some previous studies, which have used a mix of ESBL positive and negative E. coli isolates (28, 29). The association of DNA gyrase and topoisomerase IV mutations with fluoroquinolones resistance has been reported for both Gram-negative and Gram-positive organisms (12). In our study, the significant role of mutations in the resistance of E. coli to fluoroquinolones was determined. Moreover, the accumulation of mutations in gyrA and the simultaneous mutations in parC play a fundamental role in developing high level resistance to ciprofloxacin in clinical isolates (30).
The sequence analysis of QRDR in gyrA indicates that mutations occur in 2 positions at serine-83 and aspartate-87, which is similar to the report of a previous research (31). Mutations at codons 83 and 87 in gyrA are the most frequent ones among fluoroquinolone resistant E. coli. The substitution of the serine-83 by leucine is the most common change, which is consistent with the results of other studies in Iran (32, 33). It seems that this mutation is the basic change for resistance to fluoroquinolones (34). Research has shown that a point mutation in the gyrA can only slightly reduce the susceptibility of E. coli to fluoroquinolones, but high-level resistance is associated with double mutations in the GyrA protein (35). This is similar with our results indicating that a single mutation in the serine amino acid at position 83 in gyrA can only result in a low level of resistance.
Sequence analysis of QRDR of parC showed the high frequency of mutations in codon Ser80 and Glu84 among isolates. The mutation at these 2 positions has been previously reported (36). In our study, the most common mutation in parC was the replacement of serine by isoleucine at position 80, which has been known as the most common amino acid changes in the ParC protein (34). Furthermore, 22 of the isolates showed a mutation at codon 192 outside the QRDR of parC, where alanine was substituted by valine, but this substitution doesn’t apparently increase resistance to fluoroquinolones. This novel mutation needs further investigation to confirm its impact on bacterial resistance. All the isolates with mutations in QRDR of parC showed simultaneous mutations in gyrA, which is consistent with the fact that in the Gram-negative bacteria, such as E. coli, topoisomerase IV is the second target for quinolones (37). However, mutations in parC have an important role in the high level of fluoroquinolone resistance (37). It has been speculated that the high resistance to fluoroquinolones is a gradual process, which begins primarily with mutations in gyrA followed by changes in parC (38).
In conclusion, our results suggest that isolates with mutations in QRDR of both parC and gyrA have higher MIC levels compared to isolates with only mutations in gyrA. Furthermore, the highest level of resistance can be found in isolates with 2 mutations in gyrA and 2 or 3 mutations in the parC. Therefore, there is a significant correlation between the number of gyrA and parC mutations and the level of ciprofloxacin and levofloxacin resistance. These findings are in agreement with the results of other studies (16, 39). A high frequency of fluoroquinolone resistance among ESBL producing E. coli isolates in our region indicate the clustered transmission of resistance genes among ESBL producing bacteria that need regular surveillance. Consequently, the fluoroquinolones should only be used for susceptible E. coli isolates in urinary tract infections.