1. Background
Degradation of oil or petroleum by microorganisms via secretion of enzymes in the contaminated site has been produced. Bioremediation is a use of natural or genetically engineered microorganisms (GEMs) or their enzymes to clean up or neutralize oil pollutants from contaminated soil and groundwater. Bioremediation can be done in aerobic and anaerobic conditions and this technology is more economical and safer than conventional physicochemical treatments (1-3).
Oil composed mostly of diverse aliphatic and aromatic hydrocarbons and microorganisms such as hundreds of species of bacteria, archaea, and fungi can degrade it as sources of carbon and energy for growth, however, their biodegradation function on each compound may be strong and weak (4). Common bacterial species including Bacillus, Acinetobacter, Arthrobacter, Mycobacterium, Alcaligenes, Flavobacterium, and Pseudomonas have oil degradation activity (5, 6). Pseudomonas putida (P. putida) is a member of Pseudomonadaceae family and this bacterium be can used for the bioremediation process in the environment. This bacterium is a Gram-negative, flagellated, and rod-shaped that is typically found in soil and water (7, 8).
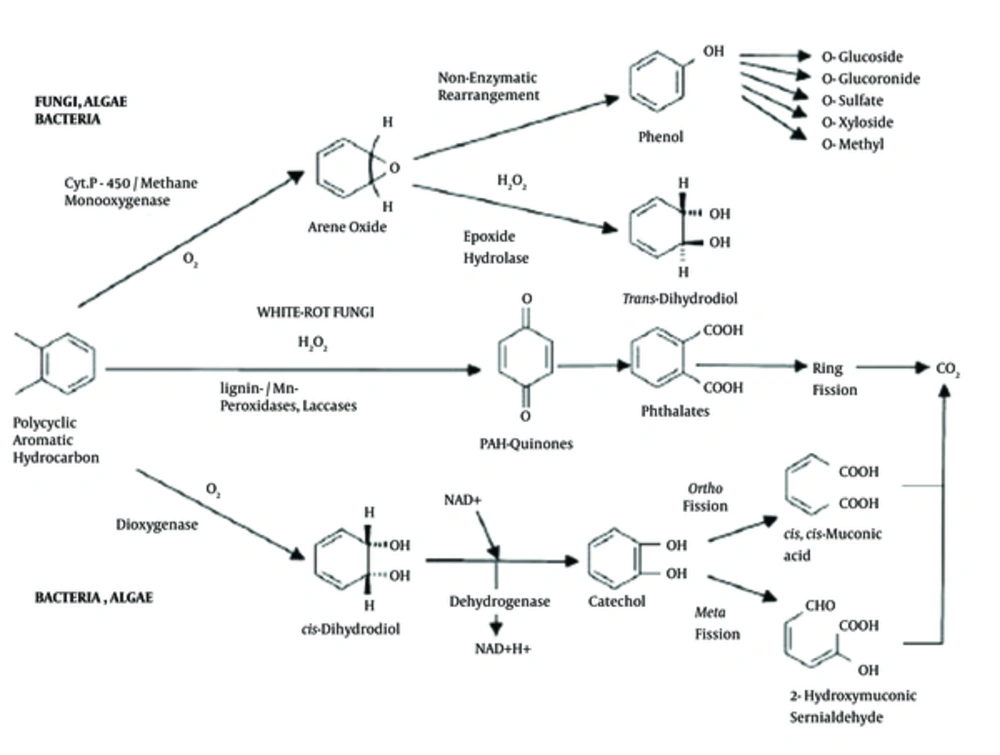
Engineering of Pseudomonas sp. by molecular techniques can be used for increasing and developing bioremediation activity of these bacteria (9). Pseudomonas sp., by producing catechol 2, 3-dioxygenase (C23O) can utilize and decomposition polycyclic aromatic hydrocarbons (PAHs). C23O is a nonheme iron dioxygenase and a key enzyme in the biodegradation of PAH compounds that is released into the environment by a large number of an industrial process by catalyzing the conversion of catechol to 2-hydroxymuconic semialdehyde (9-11).
A catechol 2,3-dioxygenase encoding gene (nahH) is one of the important genes of naphthalene catabolic plasmid NAH7 (83-kb length) in some Pseudomonas strains like P. putida and consist of 924 nucleotides that encoded catechol 2,3-dioxygenase with 36 kDa molecular weight containing 307 amino acid residues (12, 13). This plasmid helps the growth of Pseudomonas on oil- and -naphthalene contaminated environments as a sole carbon source (14, 15). The molecular degradation pathway of PAHs by dioxygenase that is encoded by the nahH gene is shown in Figure 1.
PAHs enter the environment through various routes. PAHs are ubiquitous environmental contaminants and enter the environment through various ways that are found in soil, air, water, and food sources (16, 17). PAH compounds are known as human carcinogens that spread during incomplete combustion of organic matter by industrial and other human activities (e.g., combustion of urban sewage, garbage, natural gas, vehicle traffic, aluminium smelters, tobacco smoking, cooking, power plants chemical, asphalt works, petrochemical, and oil industries) as well as natural processes like burning coal and wood or forest fires (18-21). Natural crude oil and kerosene contain significant amounts of PAHs (22). PAHs containing up to 6 fused aromatic rings and including fluoranthene, chrysene, fluorene, phenanthrene, and pyrene (17, 18).
Several PAHs in oil and petroleum products are carcinogenic, mutagenic, and teratogenic substances and associated with health hazardous and skin carcinogens (18, 23).
PAH biodegrade bacterial species that are genetically modified are useful with high efficiency for degradation of these compounds in contaminated environments by oil and petroleum products (24, 25).
2. Objectives
In this study, P. putida was genetically modified to produce dioxygenase and biodegradation capability of oil in spiked soil by this bacterium, which was evaluated by using the high-performance liquid chromatography (HPLC) method.
3. Methods
3.1. Preparation of Bacterial Strains and Culture Conditions
The present experimental study was performed from January to July 2015. The lyophilized stocks of P. putida (ATCC 12633, Persian Type Culture Collection (PTTC) No. 1694) and E. coli strain Top10F were obtained from Iranian Research organization for science and technology (IROST) and Pasteur Institute of Iran, respectively. Both bacterial stocks were suspended and cultured in the Luria-Bertani (LB) broth and incubated for 24 hours at 37°C for E. coli and 26°C for P. putida. Antibiotic susceptibility testing was performed on both bacterial strains by culturing on LB agar medium containing Ampicillin (50 to 150 mg/mL) and Kanamycin (25 mg/mL) antibiotics and showed that E. coli Top10F and P. putida strain ATCC 12633 were not resistant to these antibiotics, except Ampicillin, at concentrations 1500 X (150 mg per mL of culture medium), was suitable for P. putida transformation.
3.2. DNA Isolation
Bacterial DNA was extracted from P. putida colonies using the boiling method. The optical density of extracted bacterial DNA at a wavelength of 260/280 nm was measured by NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA) according to the method described by Sambrook and Russell, 2001 (26). The DNA samples were stored in the freezer at -20°C for further testing.
3.3. Recombinant Plasmid Preparation
In this study, pUC18 cloning and expression vector (2686 bp) with catalog number: SD0051 was purchased from Thermo Fisher Scientific Inc., Germany. Pseudomonas and especially E. coli are important hosts of this small plasmid for high copy number multiplying. The sequence of pUC18 plasmid was obtained from the Addgene site (www.addgene.org) and using DNASTAR® Lasergene software version 10 restriction enzyme patterns were identified. T/A Cloning Kit (Fermentas, Germany) was used for transferring of a plasmid into E. coli Top10F strain according to the manufacturer’s protocol.
In the present work, nahH gene synthesis and its cloning into the pUC18 vector were ordered to GeneCust Europe (Dudelange, Luxembourg) and engineered pUC18-nahH recombinant vector was generated. The accuracy of cloning of pUC18-nahH recombinant vector was found using enzymes digestion (EcoRI and BamHI restriction enzymes) and sequencing technique.
3.4. Bacterial Transformation with Recombinant Vector
For obtaining enough pUC18-nahH recombinant vector using Ins TA Clone PCR Cloning Kit (Thermo Scientific, Lithuania) this vector was transferred into E. coli Top10F according to the manufacturer’s protocol. The plasmids were extracted from bacterial cells using the Plasmid Mini Kit (BIONEER, South Korea) and after confirmation of the transferring of pUC18-nahH by HindIII enzyme digestion and PCR technique, the genetically engineered plasmid was transferred again into P. putida using the above-mentioned kit (LB media containing 150 mg per mL of Ampicillin was used for colonies selection). Then, PCR was done on purified plasmid from engineered P. putida (containing the pUC18-nahH recombinant vector) and negative control (non-engineered P. putida) to diagnose the existence of the vector containing the nahH gene.
3.5. nahH Gene Amplification
The nahH gene sequence was obtained from GenBank of National Center for Biotechnology Information (NCBI). Oligo Primer Analysis Software version 7 and Gene Runner version 3.05 were used for designing primers for gene amplification. The oligonucleotide primers sequence was compared and blasted by basic local alignment search tool (BLAST) in GenBank data and DNASTAR Lasergene software (version 10).
Polymerase chain reaction (PCR) in a total volume of 25 μL consist of 0.2 pM of each primer (NahH-F: 5´-ATGAACAAAGGTGTAATGCGC-3´ and NahH-R: 5´-TTAGGTCATAACGGTCATGAATC-3´ with Accession number: X06412), 2.5 mM MgCl2, 200 μM dNTPs mix, 2.5 μL of 10X PCR buffer (200 mM Tris-HCl (pH 8.4), 500 mM KCl), 1 unit of Taq DNA polymerase (all Fermentas, Germany), and 100 ng of isolated bacterial DNA or purified plasmid. For the no-template control (negative), 2 μL of sterile ultrapure deionised water instead of template DNA were used. The standard PCR reaction was carried out in a Gradient Palm Cycler (Corbett Research, Australia). The thermo-cycling conditions involved an initial denaturation at 94°C for 5 minutes followed by 35 cycles consisting of 50 seconds denaturation at 94°C, 50 seconds annealing at 62°C, 50 seconds of extension at 72°C, and a final extension at 72°C for 5 minutes. The PCR amplification products (6 μL/lane) and 1 kb DNA marker (Fermentas, Germany) were loaded on the 2% agarose gel electrophoresis. 1X Electrophoresis buffer (TBE): 89 mM Tris-borate, 2 mM EDTA (for 1.0 L of. 5X TBE: 54.0 g of Tris base, 27.5 g of boric acid, and 20 mL of 0.5 M EDTA,. pH 8.0). The gel was run at a constant voltage of 80 V for 30 minutes for product separation.
The gel was stained with 0.1% ethidium bromide (0.4 g/mL) and images were captured using the UVIdoc gel documentation system images (Uvitec, UK).
3.6. Soil Preparation and Characteristics for Pilot
Soil (contain sand-clay mixtures with poorly-graded gravel and clay) from the depth of 50 Cm with latitude N 31° 45ˊ08̋ and longitude E 50° 33ˊ04̋ was collected in sterile dishes from Iran. Soil compositions including sand 73%, moisture 30%, silt 15%, clay 11%, organic carbon 6.45%, gravel 1%, characterized by pH 6.9, bulk density 0.8%, uniformity coefficient (Uc) 4.1, coefficient of curvature (Cc) 0.45, and effective size 0.085 mm. In this study, both autoclaved and natural soil with microbial flora was used.
3.7. Method for Spiking Soil Samples
In the present work, soil spiking was performed based on the Brinch et al. 2002 study (27). Oil (Kerosene) was obtained from a refinery. The soil spiking procedure was carried out under sterile conditions. First, 500 g of autoclaved and natural soil were added to each dish. 125 g of soil (25% of each fraction) were spiked to oil (500 mg/kg) that were added into 100 mL acetone (Merk, Germany) as a solvent. This combined fraction was mixed completely by shacking for each dish. This mixture for evaporating the solvent was maintained at ambient temperature for 16 hours. Finally, each treatment (spiked soils) was mixed completely with the remaining 75% (375 g) of the soil sample for pilot preparation.
3.8. Soil Inoculation
Wild P. putida (non-engineered) and genetically manipulated and were cultured in 300 mL (150 mL for each dish) of LB broth medium (containing 150 mg per mL of Ampicillin antibiotic) and in 150 mL medium without antibiotic, respectively. The cultures were incubated for 24 hours at 26°C and 220 rpm in a shaking incubator. When the optical density of each cultured bacteria at 600 nm (OD600) that was measured by a 2100 visible spectrophotometer (Unico, Shanghai, China) were reached to 8 × 108 to 1.2 × 109 CFU/mL, they were used for soil inoculation. The precipitation of wild type and engineered bacteria was taken separately by centrifuging at 6000 rpm for 10 minutes. The supernatant was discarded, but 5 mL of LB medium remained for adaptation and feeding of bacteria.
Cell deposition was re-suspended in an equal volume of isotonic saline solution (containing 0.9% NaCl) and 150 mL of this solution containing bacteria was soaked to each dish. All dishes of phenanthrene and pyrene as oil indicators (500 mg/kg) were evaluated. The pilot plan including dish 1: oil (Phenanthrene + Pyrene) and Pseudomonas + autoclaved soil), dish 2 was involved soil with natural microbial flora (non-sterile) and genetically engineered P. putida (Ps. GEMs+ soil with natural microflora) to evaluate the effect of oil biodegradation of bacteria in natural conditions and dish 3 containing oil (Phenanthrene + Pyrene) Ps. GEMs + autoclaved soil.
3.9. Sampling and Extraction Method
The evaporation of phenanthrene and pyrene as an index of oil in spiked soil at the end of 3 months was 16.43% and 13.25%, respectively. Sampling and extraction were each performed in 5 days and were followed for 3 months to investigate the biodegradation of oil by engineered and wild P. putida in dishes. For sampling and extraction, 2 g of spiked soil from each dish were selected and dried at room temperature for 12 hours. Then, the soil was grinded and 2 mL acetonitrile was added. After that, oil was extracted using ultrasonic bath sonicator (CD-4820, China) for 30 minutes. Then, samples were centrifuged at 3000 rpm for 10 minutes and supernatant were followed-through from polytetrafluorethylene (PTFE) membrane filter. Finally, 20 μL of each extracted samples were injected into HPLC.
3.10. HPLC Analysis
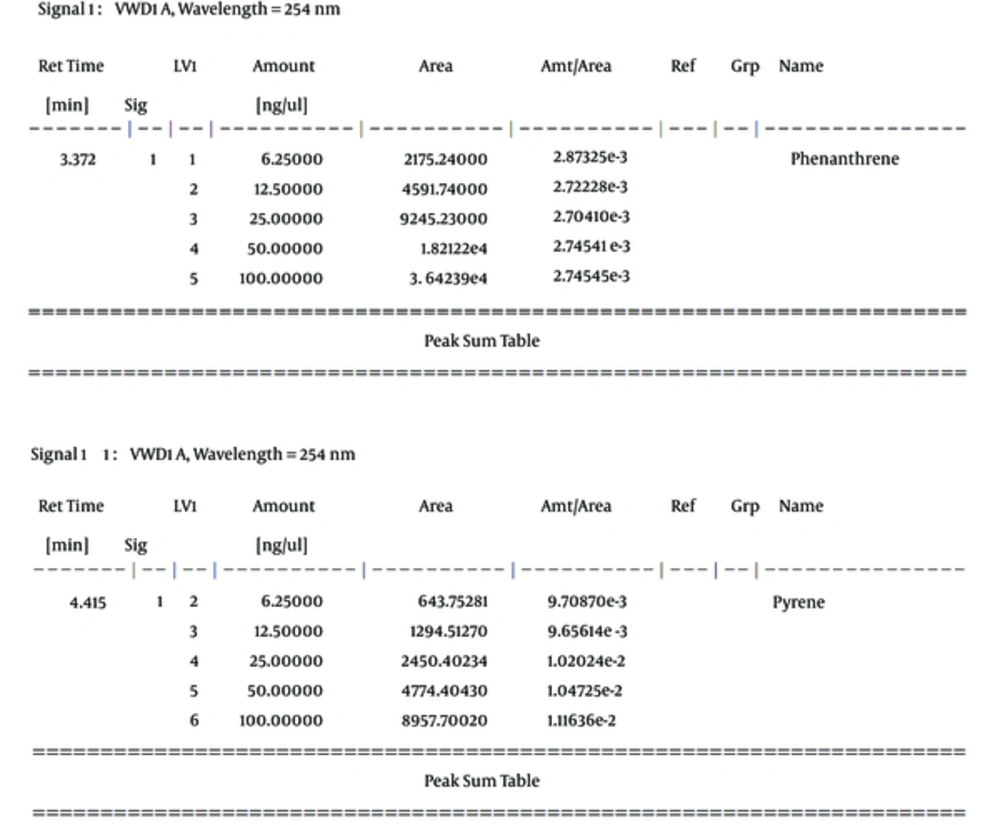
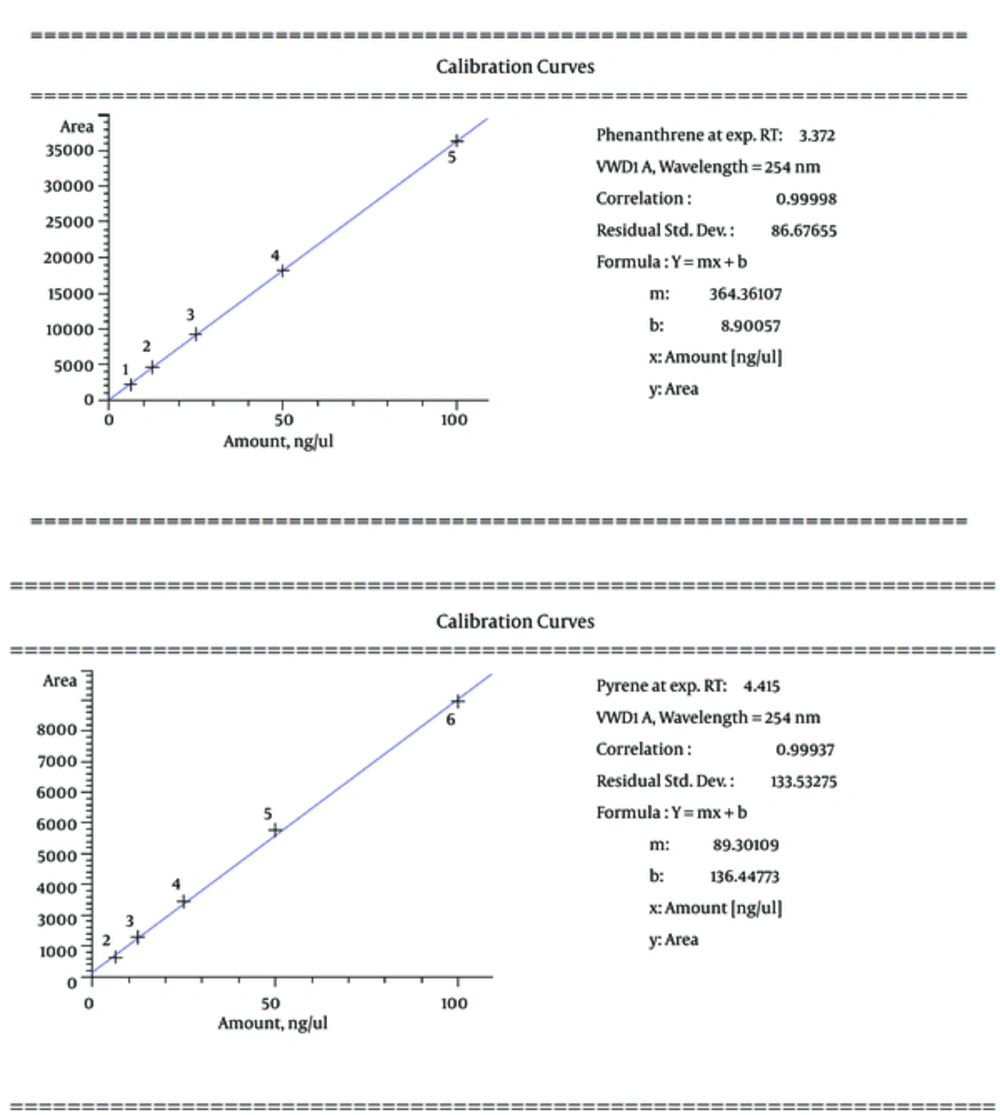
In this study, phenanthrene and pyrene as an indicator of oil were used as a serial dilution concentration (including 100, 50, 25, 12.5, and 6.25, respectively) to create standard solutions of the measurement method of HPLC (Figure 2). Then, the calibration curve for phenanthrene and pyrene was prepared with correlation coefficient equivalent 0.99998 and 0.99937, respectively (Figure 3). The mobile phase of HPLC has included 80% acetonitrile and 20% H2O with flow rate 1 mL per min, as well as the detector measured ultraviolet absorbance at 254 nm.
3.11. Confirmatory Tests
Every 30 days, 1 g of spiked soil by oil that was contaminated by wild type and engineered P. putida was mixed in sterile water injection and follow-trough from MESH filter to obtain soil extraction. Extracted soil (40 μL) were transferred on LB agar plates containing ampicillin (150 mg per mL) in sterile conditions and was cultured at 26°C for 24 hours to obtain colonies. Every 10 days, 1 g of contaminated soil were mixed in sterile saline for preparing serial dilution and were plated by drop plating for culturing on LB agar media. Plate count agar using Quebec colony counter (Bibby Sterilin Ltd, Stratfordshire, UK) was done and colony counts expressed as CFU g-1. Catalase and oxidase tests, as well as DNA extraction and nahH gene amplification by PCR, were done on each colony. Catalase test was done by mixing some colonies into a fresh drop of hydrogen peroxide (H2O2) 3% and releasing oxygen gas indicate that this test is positive. In addition, some colonies of P. putida were selected for oxidase test by transferring bacteria on filter paper that was soaked with tetra methyl-p-phenylenediamine dihydrochloride solution. Production of indophenol and purple color indicates that the oxidase test is positive.
3.12. Statistical Analysis
All experiments were performed at least 3 times. The collected data were analyzed in the Statistics programs for the Social Sciences software, version 20 (SPSS, Inc., Chicago, IL, USA). The differences between the biodegradation of oil by engineered and wild P. putida in each plate at the period of the experiment were evaluated using one-way ANOVA (Analysis of variance) followed by Tukey HSD post-hoc test for comparing the significant difference among means of various treatments. P values at or below 0.05 were considered statistically significant between groups.
4. Results
4.1. Confirmation of Cloning
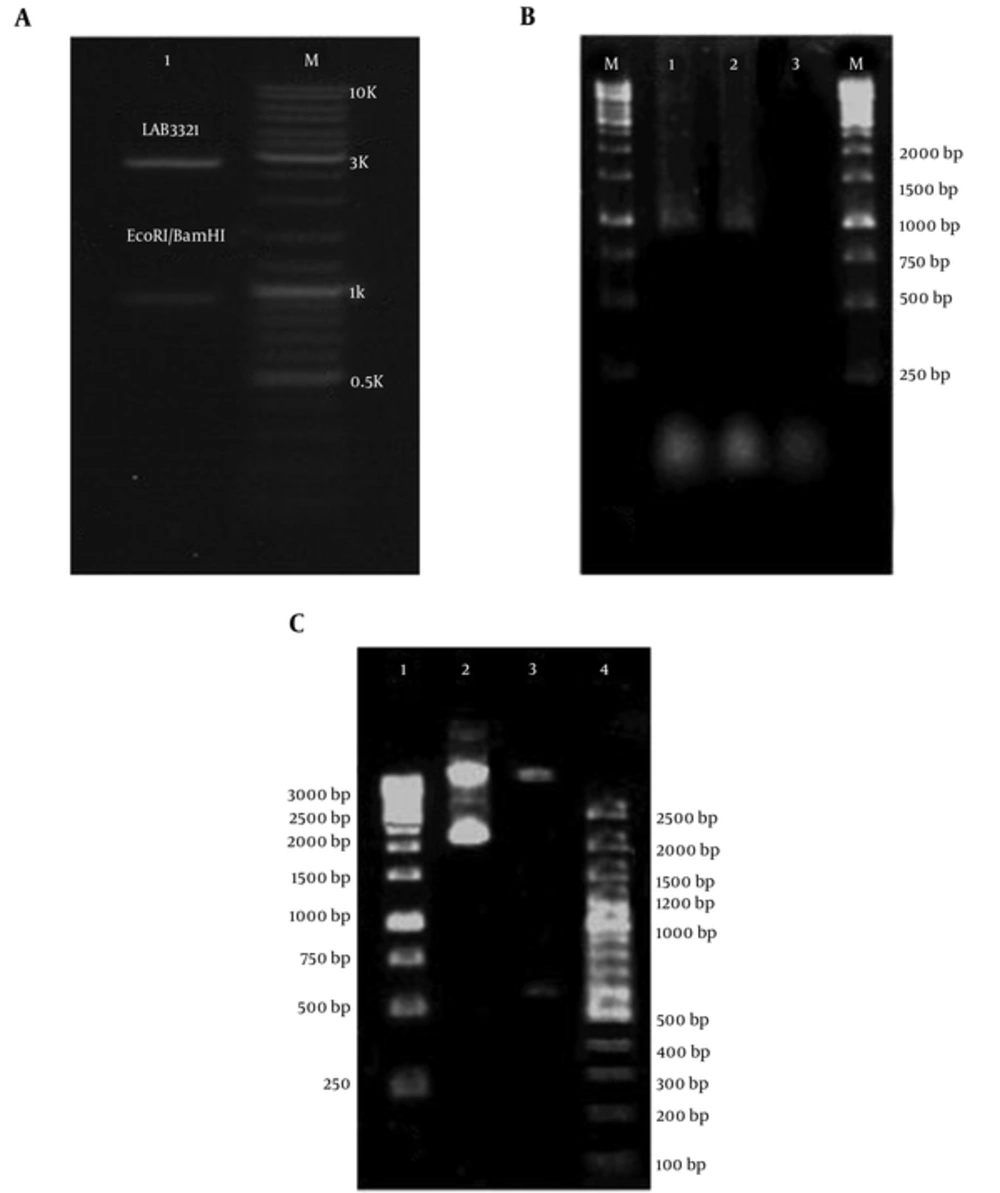
PCR technique on extracted DNA from P. putida strain (ATCC 12633) that was used in this study showed that NAH7 plasmid and nahH gene was not a presence in this bacterium. Therefore, synthesis of the nahH gene was ordered to GeneCust Europe (Dudelange, Luxembourg). BamHI and EcoRI restriction enzymes digestion and sequencing using the Sanger sequencing method were done on the engineered pUC18-nahH recombinant vector for the accuracy of nahH gene cloning in pUC18 vector and verifying the gene synthesis indicates the presence of this gene in vector (Figure 4A).
A, Digestion of pUC18-nahH Recombinant Vector Using Restriction Enzymes for Cloning Confirmation (Lane M is 100 bp Molecular Weight Markers (Fermentas, Germany), and Lane 1 is Digested pUC18-nahH Recombinant Vector (2686 and 924 bp Fragments) by BamHI and EcoRI Restriction Enzymes, Respectively); B, Amplified nahH Gene on Isolated Recombinant pUC18 Vector from Engineered E. coli (Lane M is 1 kb Molecular Ladder (Fermentas, Germany), Lanes 1 and 2 are Amplified nahH gene (924 bp), and Line 3 is Negative Control (without DNA), Respectively); C, Digestion of recombinant pUC18 Plasmid Contain nahH Gene by HindIII Restriction Enzyme for Confirmation of Transformation (Lane 1 and 4 are 1 kb and 100 bp Molecular Weight Markers (both Fermentas, Germany), Respectively, Lane 2 is Undigested Recombinant Vector (3610 bp), lane 3 is Digested Recombinant Vector (3004 and 606 bp Fragments), Respectively)
For obtaining sufficient amount of pUC18-nahH, the recombinant vector containing nahH gene was extracted from engineered E. coli. The accuracy of transformation was determined by the PCR method and HindIII enzyme digestion (were revealed 3004 and 606 bp fragments) and was confirmed the successful transformation (Figure 4B and 4C).
The pUC18- nahH recombinant vector using Ins TA Clone PCR Cloning Kit was transferred again into P. putida, successfully. Bacterial DNA was extracted from engineered and wild P. putida strains. Amplification of nahH gene (924 bp) by PCR was positive in engineered P. putida.
4.2. HPLC Data Analysis
Biodegradation of oil by wild type and genetically engineered P. putida in dishes was analyzed by the HPLC technique each in 5 days for 3 months. Data were evaluated using the one-way ANOVA followed by Tukey HSD ("Honestly Significant Difference") test for comparing the significances (Table 1). Biodegradation of oil in polluted soil by genetically manipulated P. putida (Ps.GEMs + soil with natural microbial flora and Ps.GEMs + autoclaved Soil) comparing to inoculated dish by wild-type P. putida (Pseudomonas + autoclaved soil) were statistically significant (P < 0.05). Furthermore, the statistically significance between autoclaved soil and soil contain natural microbial flora (both inoculated by genetically engineered P. putida) was not observed (P > 0.05).
| Components of Dishes | Groups | Mean Difference ± SE | Sig. P Valuea |
|---|---|---|---|
| Oil (Phenanthrene) 500 mg/kg Pseudomonas + Autoclaved Soil (1) | Oil (Phenanthrene) 500 mg/kg Ps.GEMs + Soil with Natural Microbial Flora (2) | -50.923b ± 8.373 | 0.000 |
| Oil (Phenanthrene) 500 mg/kg Ps.GEMs+ Autoclaved Soil (3) | -60.566b ± 8.373 | 0.000 | |
| Oil (Phenanthrene) 500 mg/kg Ps.GEMs + Soil with Natural Microbial Flora (2) | Oil (Phenanthrene) 500 mg/kg Pseudomonas + Autoclaved Soil (1) | 50.923b ± 8.373 | 0.000 |
| Oil (Phenanthrene) 500 mg/kg Ps.GEMs+ Autoclaved Soil (3) | -9.644 ± 8.373 | 0.487 | |
| Oil (Phenanthrene) 500 mg/kg Ps.GEMs + Autoclaved Soil (3) | Oil (Phenanthrene) 500 mg/kg Pseudomonas + Autoclaved Soil (1) | 60.566b ± 8.373 | 0.000 |
| Oil (Phenanthrene) 500 mg/kg Ps.GEMs + Soil with Natural Microbial Flora (2) | 9.644 ± 8.373 | 0.487 | |
| Oil (Pyrene) 500 mg/kg Pseudomonas + Autoclaved Soil (1) | Oil (Pyrene) 500 mg/kg Ps.GEMs + Soil with Natural Microbial Flora (2) | -43.123b ± 9.898 | 0.000 |
| Oil (Pyrene) 500 mg/kg Ps.GEMs+ Autoclaved Soil (3) | -42.342b ± 9.898 | 0.000 | |
| Oil (Pyrene) 500 mg/kg Ps.GEMs + Soil with Natural Microbial Flora (2) | Oil (Pyrene) 500 mg/kg Pseudomonas + Autoclaved Soil (1) | 43.123b ± 9.898 | 0.000 |
| Oil (Pyrene) 500 mg/kg Ps.GEMs+ Autoclaved Soil (3) | 0.78 ± 9.898 | 0.997 | |
| Oil (Pyrene) 500 mg/kg Ps.GEMs + Autoclaved Soil (3) | Oil (Pyrene) 500 mg/kg Pseudomonas + Autoclaved Soil (1) | 42.342b ± 9.898 | 0.000 |
| Oil (Pyrene) 500 mg/kg Ps.GEMs + Soil with Natural Microbial Flora (2) | -0.78 ± 9.898 | 0.997 |
Comparison the Biodegradation of oil (Phenanthrene and Pyrene as Indicators) by Wild-Type and Genetically Engineered P. putida in Dishes Using ANOVA and Tukey Post-Hoc Test
4.3. Confirmatory Tests
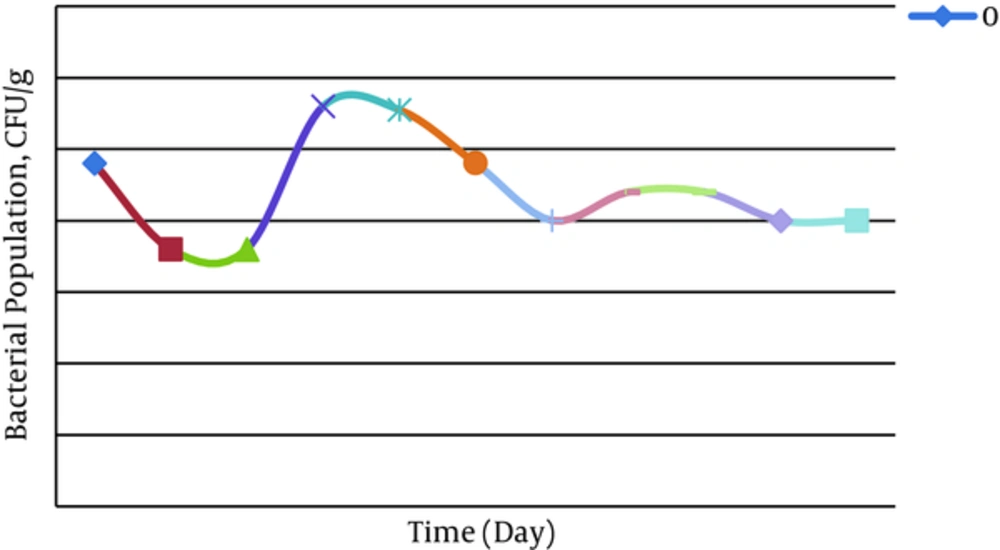
The confirmatory tests including catalase, oxidase, and gene amplification were performed on isolated bacteria from the spiked soil by oil. Catalase test on the isolated wild type and genetically engineered P. putida was positive via oxygen producing and confirmed that P. putida were active in dishes. Also, oxidase test was positive for isolated engineered P. putida from contaminated soil by oxidation of tetra-methyl-p-phenylenediamine dihydrochloride and creating a dark violet color. The amplification of C23O producing gene (nahH) in extracted DNA from isolated engineered P. putida from the spiked soil by PCR technique (924 bp) was positive and indicated that this gene was expressed in engineered P. putida. Furthermore, CFU/g, for analyzing the bacterial population in spiked soil by oil, is shown in Figure 5.
Evaluation of Isolated Bacterial Population (CFU/g) from Polluted Soil by Oil Was Done Every 10 Days (In 0 - 20 Days After Inoculation, P. putida Population Were Decreased and It may be Related to Adaptation of Bacteria in New Environment, 20 - 40 Days After Inoculation CFU was Increased and Then (40 - 60 Days) the Population Were Decreased Again, and finally the Bacterial Population Close to Stationary Phase).
5. Discussion
Application of genetically engineered bacteria (bioremediation) improved biodegradation of pollutants in environments (28). In this study, genetically engineered P. putida that produce dioxygenase enzyme was generated via transferring of pUC18-nahH recombinant vector and were used for biodegradation of oil in the spiked soil. Removing of oil from the soil by genetically manipulated and wild P. putida was evaluated using the HPLC measurement method. The results indicated that biodegradation of oil from spiked soil using genetically manipulated P. putida were statistically significant (P < 0.05) comparing to soil contaminated by a wild type of P. putida. Also, biodegradation activity of genetically engineered P. putida between autoclaved soil and dish that contain natural microbial flora were not statistically significant (P > 0.05). Viability and activity of isolated engineered P. putida from the spiked soil with oil by culturing, as well as catalase, oxidase, and PCR tests were investigated and explanatory degradation of oil via these bacteria by nahH gene expressing in the soil environment.
A number of studies have been performed for biodegradation of PAH compounds by Pseudomonas and other gram-negative bacteria as well as fungi such as Trichoderma/Hypocrea genus; however, no studies to date have been performed on the degradation of oil (kerosene) in soil by genetically engineering P. putida, that produce the nahH gene. In one study naturally adapted P. putida isolated from oil-contaminated sites in India was isolated and evaluated for biodegradation of crude oil in the open environment (29, 30). In another study, the efficiency of Bacillus subtilis DM-04 and Pseudomonas aeruginosa M and NM strains are isolated from a petroleum-contaminated soil sample from North-East India on biodegradation of crude petroleum-oil hydrocarbons in soil and shake flask compared and indicated P. aeruginosa strains were more efficient than B. subtilis strain and as well as showed B. subtilis DM-04 and P. aeruginosa M and NM strains could be effective for in situ bioremediation (31). The application of P. putida ZWL73 in contaminated soil with 4-chloronitoben-zene (4CNB) was investigated by Niu et al. (32). In another research, it was found that tfdA gene located in plasmid pRO103 in phenol-degrading recipient strains leads to significantly biodegradation rate of phenoxy acetic acid in sterile and non-sterile soils (33). In the present study, the pUC18-nahH recombinant vector was generated and transferred into P. putida and degradation of oil in polluted soil was increased statistically via evaluation of phenanthrene and pyrene as an indicator. In another study by Cao et al. (2012), P. putida KT2440-rhlABRI was genetically generated by cloning and transformation of rhlABRI cassette gene of P. aeruginosa BSFD5 and indicated that this manipulated strain could increase the dissolution of pyrene by promoting its degradation by indigenous microorganisms in soil (34). While in our study encoded C23O enzyme gene (nahH) was cloned and pUC18-nahH was generated for transferring into P. putida and used for removing of oil from the soil. In the study of Zhou et al. (2013), the C23O gene was cloned into the plasmid pK4 derived from pRK415 and transferred into Pseudomonas sp. CGMCC2953, successfully. The capability of this modified strain was investigated by increasing of phenanthrene decomposition (35). In the present study, the pUC18-nahH recombinant vector was successfully created and transferred into P. putida. The biodegradation activity of engineered P. putida in spiked soil (autoclaved soil and natural soil containing microbial flora) on oil biodegradation showed oil removal compared to wild-type group was increased statistically significant. In the recent study by Samin et al. 2014 genetically engineered P. putida via cloning of dehalogenase gene (dhaA31) generated for evaluation the 1,2,3-Trichloropropane (TCP) degradation activity (36). Same to the findings of the present study PAH compound in oil and chlorinated hydrocarbon can effectively biocatalyst by GEMs.
Biodegradation of major oil and PAHs pollutions in soil and water could be more helpful for decreasing of their affects on the environment and its exposures and associated health risks in human. The genetically manipulated P. putida containing a recombinant pUC18-nahH vector that was produced in this study can effectively biocatalyst and remove oil from spiked soil as well as be used as biodegradable strain in environmental and industrial pollutions. As a suggestion, in future work it would be better if P. putida were genetically manipulated for other genes like nahA and nahE that contribute to PAHs degradation.