1. Background
The gastrointestinal tract (GIT) is colonized with a complex and dynamic microbial community known as gut microbiota (1). The gut microbiota plays significant role in human health and diseases (2). It has important effects on host functions, including homeostasis of the GI, metabolism, immune, and nervous system (3). This complex community is composed of diverse microorganisms dominated by bacteria. Most of these bacterial species belong to phyla Firmicutes and Bacteroidetes. Moreover, other phyla such as Verrucomicrobia and Actinobacteria are present in gut microbiota (4). The composition of gut microbiota is established during 2 - 3 first years of life under control of many factors, including mode of delivery, genetic background, geography, nutrition, physical activity, and gender (5).
In normal conditions, symbiosis relationship between gut microbiota and host, the gut microbiota-host interaction is balanced. Conversely, in dysbiosis, alternation of gut microbiota pattern, putative interaction is disturbed. Dysbiosis resulted from high fat diet (HFD) induces low-grade inflammation, which leads to insulin resistance (IR). This condition is considered as turning point of various disorders and diseases such as obesity, metabolic syndrome, type 2 diabetes, inflammatory diseases (inflammatory bowel disease), and colorectal cancer (CRC) (6, 7).
Recent studies consider the gut microbiota as an environmental factor that has important role in the pathophysiology of obesity (8). Energy harvest increase changes in microbial components and metabolites are attributed to the alternation of gut microbiota composition in obesity. These events are parallel with increase of Firmicutes to Bacteroidetes ratio in obese subjects (9, 10). Therefore, this study aimed to investigate the gut microbiota composition.
It seems that Bacteroides spp. among the gut microbiota members has an important role in pathophysiology of obesity. Bacteroidetes are Gram-negative rods, anaerobic and non-spore-forming bacteria. Bacteroidetes spp. extract energy from protein and carbohydrates by fermentation (11). High enzymatic potentials of B. fragilis and B. thetaiotaomicron contribute in maintenance of homeostasis (12). Also, these bacteria have immunomodulatory effects that induce tolerance to gut microbiota (13). Hence, due to metabolic and immune potentials of B. fragilis and B. thetaiotaomicron, their frequency could be important in obesity treatment. Population criteria (genetic background, ethnicity, diet, lifestyle, geography distribution) could affect the gut microbiota pattern. Accordingly, in this study, for the first time, we focused on B. fragilis and B. thetaiotaomicron relative abundance in Iranian population.
2. Methods
2.1. Study Population
A total of 100 healthy adults (age range: 20 - 60 years) were recruited. These subjects were assigned into two equal groups of normal and obese based on body mass index (BMI). Subjects with BMI between 18.5 and 24.9 kg/m2 were considered as normal group. Participants with significant infection, chronic diseases, and the use of antibiotics and corticosteroids were excluded. This study was performed in accordance with the ethical rules of the Helsinki Declaration.
2.2. DNA Extraction from Stool Samples
The fresh stool samples were collected from subjects and stored immediately at -20°C. DNA was extracted using a QIAamp DNA stool mini kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The quality of extracted DNA was assayed by agarose gel electrophoresis and spectrophotometric analysis. All DNA samples were stored at -20°C.
2.3. Quantitative Polymerase Chain Reaction (qPCR)
The abundance of bacteria was estimated by universal primers, which amplified a conserved region of the 16S rRNA gene. SYBR Green qPCR was conducted using Light Cycler® 96 SW 1.1 (Rocsh, Germany). Each reaction mixture of 20 μL was composed of SYBR Premix Ex Tag II (RR820L-Takara), 0.5 μL of each of the specific primers, and 5 μL of template DNA. The amplification program was designed according to appropriate annealing temperature consisting of 1 cycle of 95°C for 60 s, followed by 40 cycles of denaturation at 95°C for 5 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s (14).
2.3.1. Standard Curve
The abundance of bacteria was calculated through DNA concentration. For this purpose, serial dilutions of DNA from standard strain Escherichia coli were prepared. The standard curve allows to calculate DNA concentration of each bacteria in stool samples (15).
2.4. Statistical Analyses
In this study, categorical variables are presented as number (percent) and continuous variables as mean ± SD. Independent t-test was used to assess mean differences between normal and obese groups. Statistical analyses were performed using SPSS software version 21.0 (SPSS Inc., Chicago, IL, USA). All statistical tests were 2-tailed, and a P ≤ 0.05 was considered statistically significant.
3. Results
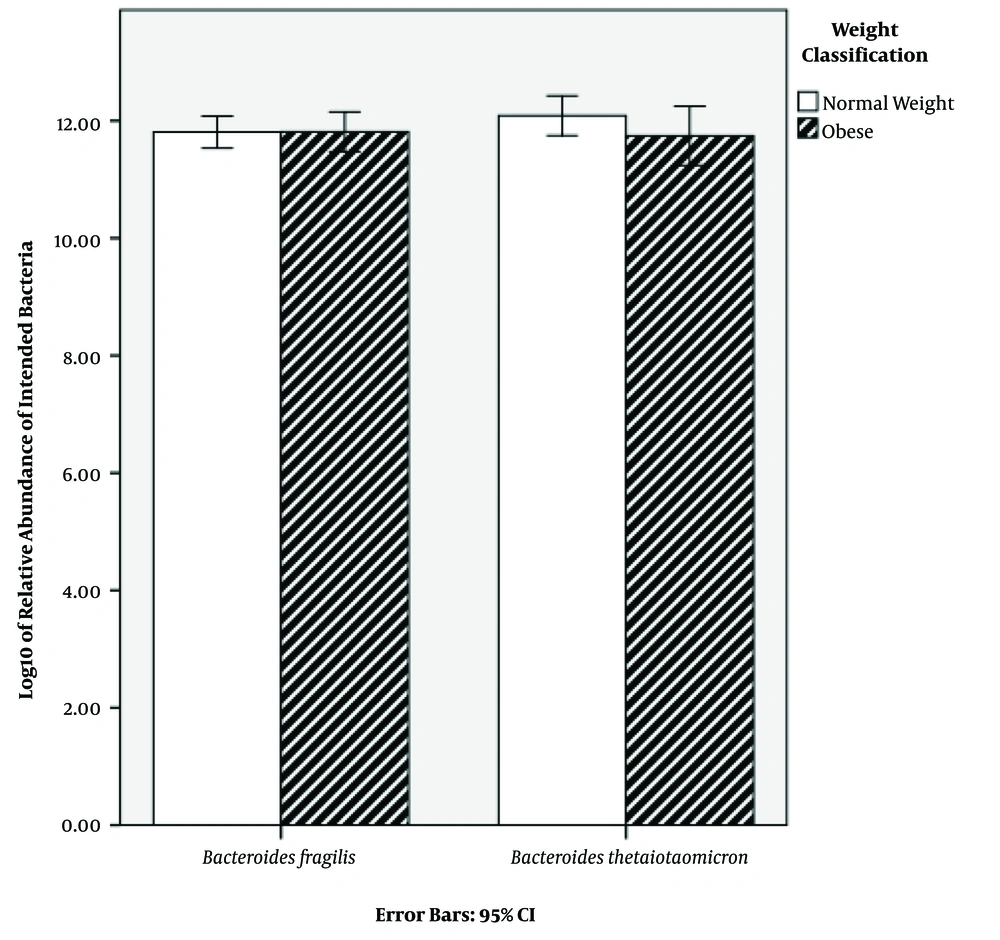
Characteristics of the subjects are shown in Table 1. BMI classification consisted of two groups: normal weight (50%) and obese (50%) (Table 1). The results of qPCR showed that the relative abundance mean of B. fragilis in normal weight and obese subjects was 8.68 × 1012 and 9.27 × 1012 CFU mL-1, respectively. Additionally, the relative abundance mean of B. thetaiotaomicron was 2.32 × 1012 and 5.39 × 1012 CFU mL-1 in normal weight and obese subjects, respectively (Table 2). In order to find a correlation between B. fragilis and B. thetaiotaomicron abundance and obesity, the bacterial concentrations were analyzed between normal and obese subjects. Although the obese subjects had more B. fragilis and B. thetaiotaomicron abundance compared to subjects with normal weight, no significant difference was identified between relative abundance of B. fragilis (P = 0.79) and B. thetaiotaomicron (P = 0.18) in the two groups (Figure 1).
| Obese | Normal-weight | |
|---|---|---|
| Subjects (male/female) | 26 (12/14) | 28 (12/16) |
| Age (y) | 43.56 ± 1.92 | 30.55 ± 1.42 |
| Weight, kg | 81.38 ± 2.74 | 63 ± 1.32 |
| Height, m | 1.68 ± 0.025 | 1.68 ± 0.010 |
| BMI, kg/m2 | 28.42 ± 0.59 | 22 ± 0.36 |
| BMI s.d. score | 3.01 | 1.89 |
| Relative Abundance of B. thetaiotaomicron | Relative Abundance of B. fragilis | |
|---|---|---|
| Mean | 3.8611×1012 | 8.9777×1012 |
| Std. error of mean | 1.16127×1012 | 1.12594×1012 |
| Std. deviation | 1.16127×1013 | 1.12594×1013 |
| Minimum | 5.80×105 | 9.22×107 |
| Maximum | 7.75×1013 | 5.20×1013 |
4. Discussion
The gut microbiota is a diverse and complex microbial community which affects metabolism and energy homeostasis. The gut microbiota composition is associated with health and diseases (3). The pattern of gut microbiota has been established during the first 2-3 years of life and affects many aspects in the host, including immune cell functions, glucose and lipid metabolism, energy homeostasis, and susceptibility to disease (16, 17). The alteration of gut microbiota could be spotted as a biomarker in several disorders and diseases such as obesity (18-21). Bacteroidets has significant roles in gut microbiota-host interaction. Therefore, the determination of abundance could be valuable in controlling obesity, in this study, for the first time, the relative abundance of B. fragilis and B. thetaiotaomicron was determined in Iranian population.
Recently, a correlation between gut microbiota composition and obesity was identified (22-24). Various studies have reported that obese and non-obese subjects have different gut microbial compositions (14). Bacteroidets and Frimicutes are two important phyla that constitute gut microbial community. Bervoets et al. showed an elevated Firmicutes to Bacteroidetes ratio in obese children (14). B. fragilis and B. thetaiotaomicron are anaerobic intestinal commensal that belong to Bacteroidetes phylum. They have significant metabolic and immune potentials in gut microbiota-host interaction (13). Hence, their relative abundance could influence the host's metabolism.
There are various reports about abundance of B. fragilis and B. thetaiotaomicron. For example, Vael et al. proved that high intestinal B. fragilis abundance in infants between the age of 3 weeks and 1 year was associated with a higher risk of obesity in their adulthood (25). Karlsson et al. did not observe a significant difference between B. fragilis abundance in obese children compared to normal-weight subjects (26).
Kasai et al. demonstrated greater bacterial diversity and different gut microbial composition in obese compared with non-obese subjects. Also, their results showed that some bacterial species, including B. thetaiotaomicron, had significantly greater concentration in the non-obese group (27).
The establishment of gut microbiota composition is influenced by various factors including, mode of delivery, genetic background, lifestyle, geography, ethnicity, diet, etc. Therefore, these factors affect the results of gut microbiota and obesity studies.
4.1. Conclusion
Although the obese Iranian subjects had more fecal B. fragilis and B. thetaiotaomicron compared with normal-weight subjects, no significant difference was identified between the relative abundance of B. fragilis and B. thetaiotaomicron in obese and non-obese individuals. Since B. fragilis and B. thetaiotaomicron have important roles in metabolism and energy homeostasis, determining the bacterial relative abundance could be valuable in controlling obesity. In total, determination of gut microbiota pattern is essential regarding introducing some effective prevention and treatment strategies in obesity as well as other disorders.