1. Background
The liver is the primary metabolism site in the body that is usually subject to a high degree of exposure to medicines and external substances (1). Therefore, drug-induced liver injury is the most important clinical problem that accounts for about 10% of the liver transplant cases and may lead to patient death (2). Acetaminophen is the most common drug metabolized by the liver and some of it at high doses is converted into the active metabolite of N-acetyl-p-benzoquinone imine (NAPQI) by the cytochrome P450 system, causing acute liver failure (3). The measurement of serum alanine aminotransferase (ALT) is a standard biomarker for the diagnosis and prognosis of liver injuries (4) that is released into the bloodstream due to injury to the cell membrane (5). However, ALT is not liver tissue-specific and can be released by injured cells of other organs such as kidneys, muscles, and heart (5).
An ideal biomarker should have low concentrations in the blood and other body fluids, and can be released into the circulatory system and measured quickly and accurately. MicroRNAs are noncoding and single-stranded molecules of intracellular regulating RNAs with 18 - 25 nucleotides in length (6) that adjust gene expression through the RNA interference (RNAi = RNA interference). miRNAs exist in a variety of body fluids, including blood (7), urine (8), and saliva (4) and control important processes such as development, differentiation, proliferation, and apoptosis (9). In addition to their role in regulating gene expression, serum miRNA levels are recently introduced as a new class of biomarkers for various diseases such as fibrosis (10), heart failure (11), and cancer (12). Many miRNAs are expressed in tissues and some of them show a high tissue specificity (13) such as miR-122, the most abundant miRNA in the liver (14); it is a multi-functional RNA (6) playing an important role in oxidative stress pathways (15) and metabolism of lipids (14). Although miR-122 is specific to liver tissue, it exists in small amounts in the heart tissue. Recent studies have shown that high serum levels of miR-122 are associated with a range of liver diseases such as fibrosis, cancer, or viral infections.
Since miR-122 has a high specificity and is non-invasive, it is considered a good biomarker for liver diseases (5). miR-192, the other abundant miRNA in the liver (6), is a general gene expression regulator that helps TP53 suppressor function in the liver injury and has an anti-apoptosis role. In a previous study, Sanchari Roy showed that an increase in the serum level of miR-192 in mice indicated the liver injury. Considering that the level of alanine transaminase as the biomarker of liver injury is not detectable at least up to 12 hours after the ingestion of high doses of acetaminophen and since this time is very important for the development of liver injury (16).
2. Objectives
The aim of this study was to evaluate the use of miRNAs as reliable and suitable biomarkers for diagnosing acetaminophen-induced liver injuries, in comparison with the histopathology and enzyme assay.
3. Methods
3.1. Chemicals
APAP was purchased from the MERCK Company (St. Louis, USA). MicroRNA extraction was carried out using the miRNeasy Serum/Plasma kit (Qiagen, USA) while the mir-amp kit from Parsgenome Company (Iran) was used for miRNA amplification.
3.2. Animal and Care Conditions
A total of 32 white male Sprague-Dawley rats aged 6 to 32 weeks and weighing 180 g were randomly selected from the Laboratory Animal Research Center of Mazandaran University of Medical Sciences. The animals were kept in cages made of transparent polycarbonate at the appropriate temperature and humidity. They had adequate access to food and water. All test steps were performed according to the guidelines issued by the Ethics Committee for working with Laboratory Animals of Mazandaran University of Medical Sciences. The rats were later divided into eight groups (four rats each) and kept under similar conditions. Three groups were sampled in the first hour of injection and three other groups three hours after the injection.
Toxic and therapeutic doses (75, 150, and 300 mg/Kg b.w) of acetaminophen were intraperitoneally injected to each rat in the experimental groups at a time. Phosphate buffered saline (PBS) (100 mM, pH = 7.40) was injected only into the two groups, each of which was used as a control for the first-hour and third-hour injections.
3.3. Blood Chemistry and Histopathology
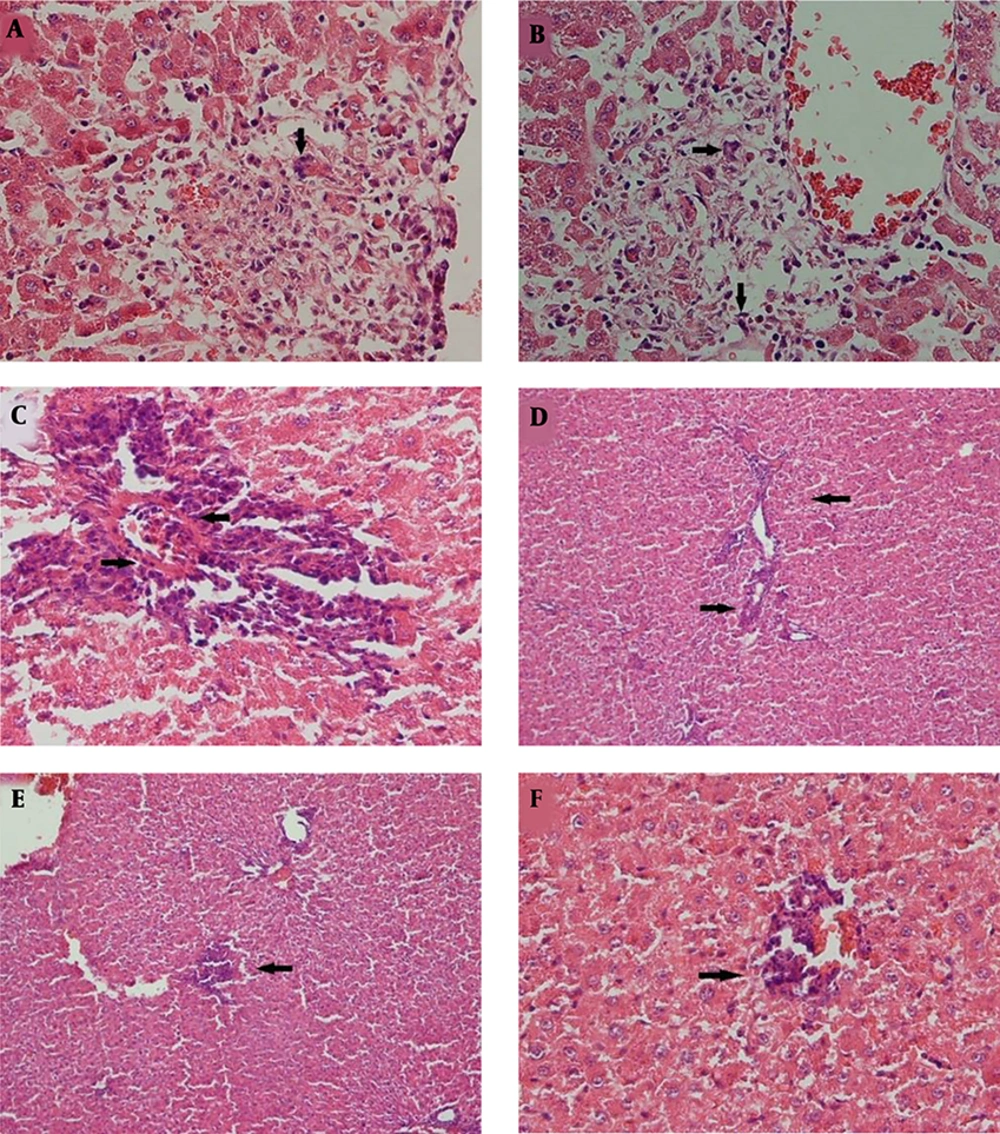
After inducing full anesthesia using ether, blood samples were taken from the heart (cardiac puncture). Serum ALT and AST were measured by a Hitachi 911 autoanalyzer (Hitachi High-Technologies, Japan). After recording the macroscopic tissue changes, the liver of the tested rats was fixed in 10% formalin. After routine pathological processing, five micrometer-thick sections were prepared and stained with hematoxylin and eosin.
To investigate four hepatocyte necrosis factors, inflammation of the portal region, inflammatory cell infiltration, necrosis of centrilobular area, and the reduction of cellular glycogen were used. The severity of hepatic injury rated from 0 to 3 (0 = no injury; 1 = mild injury; 2 = moderate injury; and 3 = severe injury) was studied and evaluated (17).
3.4. microRNA Extraction
The extraction process was carried out using kit column miRNeasy Serum/Plasma kit (Qiagen, USA) so that the first 200-μL volume of plasma samples was mixed with 1000 μL of the QiAzol solution and the process of microRNA extraction continued according to the manufacturer's kit protocol. Finally, the product resulting from the extraction was kept in a freezer at -80°C for later use.
3.5. cDNA Synthesis and Real-Time PCR
First, a polyA tail was added to the microRNA. To do so, the Parsgenome miR-Amp kit (Parsgenome Company, Iran) was used according to the manufacturer's protocol. Then, 2 μL of the miRNA polyA tail product was used to make 20 μL cDNA. The cDNA was made using Parsgenome MiR-Amp universal cDNA synthesis kit according to the manufacturer’s instruction.
The miRNA levels were measured by fluorescence signals generated by an SYBR Green probe during the real-time PCR in Bio-Rad IQ5 device (Bio-Rad, USA). 1.3 μL of specific primers were used to amplify cDNA in a total volume of 20 μL reactions using the Parsgenome miR-Amp kit. Finally, the amount of expression of each miRNA was calculated using the Bio-Rad LQ5 software provided by the company (Bio-Rad, USA). Moreover, 5s rRNA was used as an internal control to study hepatic microRNAs in rats. It has been shown that 5s rRNA has a regular and stable expression in plasma samples (18).
3.6. Statistical Analysis
Quantitative data were expressed as means ± SD. Box-Cox transformation was used to normalize data due to non-homogeneity of variances and the parametric tests were used accordingly. In addition, the correlation of the data was evaluated using the multivariate analysis of variance (MANOVA). The data were analyzed using SPSS V.21.3.0.2 and the P value < 0.05 was considered as a significant criterion in all the tests. GraphPad Prism V. 6 software was used for diagramming. The ΔCt method was used to determine the relative expression of miRNA levels, which was later normalized using 5S rRNA. The data were reported as 2-ΔCt using the following formula: ΔCt = Ct (miRNA of interest) - Ct (housekeeping). Finally, the level of change was expressed as 2 [- (mean of ΔCt values of treated samples - mean of ΔCt values of control samples)] (19).
4. Results
4.1. Histopathological Analysis
Histopathological data of this study have been reported recently (17). Briefly, the histopathological changes were seen in the liver of rats with 150 mg acetaminophen injection in both one and three-hour tests in the form of hyperemia, mild edema in the portal area, and mild infiltration of inflammatory cells. At one-hour and three-hour injections of 300 mg acetaminophen, mild centrilobular necrosis was seen, but the spotty necrosis of liver cells was slightly more in one hour than in three hours of 300 mg injections (Table 1) (17).
| Pathology State/Groups | Control | 75 | 150 | 300 | ||||
|---|---|---|---|---|---|---|---|---|
| 1h | 3h | 1h | 3h | 1h | 3h | 1h | 3h | |
| Portal area inflammation and hematoma | ||||||||
| Grade 0 | + | + | + | + | ||||
| Grade 1 | + | + | + | + | ||||
| Grade 2 | ||||||||
| Grade 3 | ||||||||
| Liver cell necrosis (spotty necrosis) | ||||||||
| Grade 0 | + | + | + | + | + | + | ||
| Grade 1 | + | |||||||
| Grade 2 | + | |||||||
| Grade 3 | ||||||||
| Infiltration of inflammatory cells | ||||||||
| Grade 0 | + | + | + | + | ||||
| Grade 1 | + | + | + | |||||
| Grade 2 | + | |||||||
| Grade 3 | ||||||||
| Centrilobular necrosis and glycogen decrease in cell | ||||||||
| Grade 0 | + | + | + | + | + | + | ||
| Grade 1 | + | + | ||||||
| Grade 2 | ||||||||
| Grade 3 | ||||||||
a +, The level of changes detected in each group.
4.2. ALT and AST Levels in Blood
The liver enzyme activity of this study has been reported recently (17). Briefly, AST or ALT blood levels were measured in all serum samples. Based on the multivariate analysis of variance (MANOVA), both main effects (dose and time) and their interactions were significant (dose: Wilks’ λ = 0.228, P < 0.001, time: Wilks’ λ = 0.456, P < 0.001, and their interaction: Wilks’ λ = 0.493, P = 0.010). In other words, the rates of ALT activity (IU/L) at the dose of ≥ 75 IU/L at the first and third hours were significantly different from each other (dose 75 IU/L: P < 0.002, dose 150 IU/L: P = 0.025, and dose 300 IU/L: P < 0.001) (Table 2) (17).
| Time, h | Dosage, mg/kg | AST (Mean ± SD) | ALT (Mean ± SD) |
|---|---|---|---|
| 1 | Control | 183.5 ± 6.5 | 70.5 ± 7 |
| 75 | 307.3 ± 138 | 70.5 ± 8 | |
| 150 | 227.5 ± 40 | 90.0 ± 8 | |
| 300 | 251.2 ± 75 | 78.0 ± 7 | |
| 3 | Control | 187.0 ± 8 | 68.2 ± 10 |
| 75 | 365.5 ± 156 | 143.2 ± 88 | |
| 150 | 293.2 ± 124 | 151.5 ± 65 | |
| 300 | 347.5 ± 135 | 162.5 ± 35 |
4.3. MiRNAs Level in Plasma
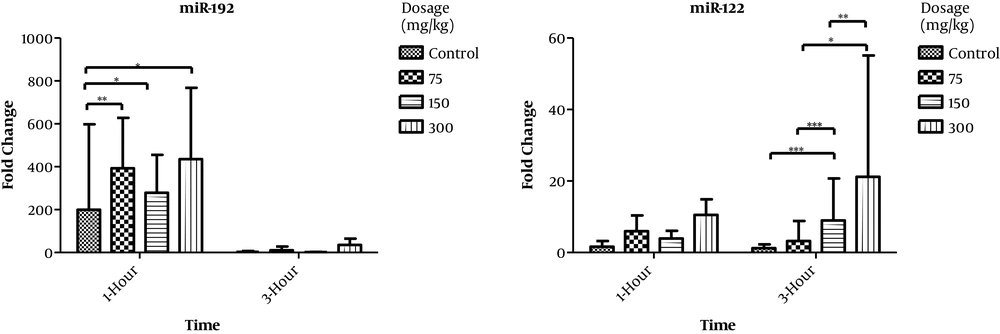
The levels of miR-122 and miR-192, as well as 5s rRNA as a control, were examined in the plasma of rat model with acute liver injury using real-time PCR. The amounts of miR-122 were increasing at 75, 150, and 300 mg/kg doses both at one and at three hours after injection. The observations showed that there was a statistically significant correlation between miR-122 and miR-192 (R = 0.604 and P < 0.001). Moreover, the present study revealed that there was a partial correlation between miR-122 and miR-192 (R = 0.536 and P < 0.003) when the effect of dose and time was controlled (Table 3 and Figure 2).
| Case Summaries | |||||
|---|---|---|---|---|---|
| Time, h | Dosage, mg/kg | miR-122 | miR-192 | ||
| N | Mean ± SD | N | Mean ± SD | ||
| 1 | 0 | 4 | 1.63 ± 1.62 | 4 | 199.53 ± 398.74 |
| 75 | 4 | 5.96 ± 4.41 | 4 | 393.1 ± 234.68 | |
| 150 | 4 | 3.9 ± 2.18 | 4 | 278.46 ± 177.21 | |
| 300 | 4 | 10.51 ± 4.37 | 4 | 411.93 ± 275.64 | |
| Total | 16 | 5.5 ± 4.54 | 16 | 320.75 ± 268.88 | |
| 3 | 0 | 4 | 1.25 ± 1.02 | 4 | 2.89 ± 4.52 |
| 75 | 4 | 3.24 ± 5.6 | 4 | 10.64 ± 17.12 | |
| 150 | 4 | 183.45 ± 6.71 | 4 | 1.82 ± 1.37 | |
| 300 | 4 | 21.18 ± 33.94 | 4 | 26.8 ± 29.06 | |
| Total | 16 | 52.28 ± 80.17 | 16 | 10.54 ± 18.37 | |
| Total | 0 | 8 | 1.44 ± 1.27 | 8 | 101.21 ± 281.42 |
| 75 | 8 | 4.6 ± 4.89 | 8 | 201.87 ± 255.97 | |
| 150 | 8 | 93.68 ± 96.08 | 8 | 140.14 ± 187.95 | |
| 300 | 8 | 15.85 ± 23.12 | 8 | 219.37 ± 274.41 | |
| Total | 32 | 28.9 ± 60.7 | 32 | 165.65 ± 244.91 | |
Changes of miR-122 and miR-192 levels in blood plasma of experimental rats. The amounts of miR-122 are increasing with 75, 150, and 300 mg/kg doses, both at one and at three hours after injection. The amounts of miR-192 are increasing after one hour of APAP injection, but not after three hours.
The results of multivariate analysis of variance (MANOVA) and univariate analysis of variance are reported in Table 4. MANOVA showed the effects of time, dose, and their interaction were statistically significant. Wilks' Λ showed that 28% of the variance of miR-122 and miR-192 was not explained by the level of interaction between time and dose (Wilks' Λ = 0.28, P < 0.001). According to univariate analysis, the significance of the interaction term that was shown by the multivariate test was related to miR-122. Based on the result, 53.6% of the variance of miR-122 was explained by the level of interaction between time and dose (Partial η2 = 0.536, P < 0.001).
| Factors | Multivariate | Univariate | ||||
|---|---|---|---|---|---|---|
| Wilks' Λ | P Value | miR-192 | miR-122 | |||
| Partial η2 | P Value | Partial η2 | P Value | |||
| Time, h | 0.421 | < 0.001 | 0.451 | < 0.001 | 0.032 | 0.378 |
| Dose, mg/kg | 0.274 | < 0.001 | 0.357 | 0.013 | 0.605 | < 0.001 |
| The interaction between time and dose | 0.280 | < 0.001 | 0.252 | 0.068 | 0.536 | < 0.001 |
a The result has been computed under the natural logarithm transformation.
5. Discussion
The prevalence of drug-induced liver injury has been reported over 10 to 15 cases per 100000 individuals per year (20). To date, only a few metabolites or enzymes have been used to assess the drug-induced liver injury. These tests include the determination of total serum bilirubin levels, activity of alkaline phosphatase, aspartate aminotransferase, and alanine transferase enzymes (21). Increases in these enzymes are usually related to biles and hepatocytes damages, but they are not specific only to liver damage. Although the ALT activity is generally susceptible to liver damage, it is not susceptible to time or kinetics (22). As a result, finding new biomarkers is important for early diagnosis and treatment. The present study investigated miR-122 and miR-192 as the biomarkers for APAP-induced liver injury in Sprague-Dawley rats at different doses of 75, 150, and 300 mg/kg in the first and third hours, compared to liver enzymes and pathological observations. In the first hour of APAP drug administration, no particular changes in ALT enzyme activity were seen at different doses compared to the normal control. However, two folds of changes at ALT level were observed when it was evaluated after the third hour (17). ALT and AST levels rise during various diseases like kidney damage; so, they cannot be considered as specific biomarkers for liver injury.
The hepatotoxicity rat model used in the present study showed that miR-122 and miR-192 plasma levels increased due to liver injuries. miR-122 is a known biomarker for liver toxicity in rats (23), monkey (1), and humans (16, 24). The overexpression of miR-122 increases the level of P53, an important suppressor gene that causes apoptosis by regulating the PI3K (phosphoinositide 3-kinase)/Akt signaling pathway (25). Previous studies proposed miR-122 and miR-192 as good candidate biomarkers that their levels increase both in plasma and in the liver during the drug-induced liver injury (26-28). MiR-122 was reported as the most abundant liver-specific miRNA, comprising about 70% of total hepatic microRNA (29). In a study, Wang et al. evaluated a total of 93 different menus from the liver and plasma in mice. Several microRNAs expressed in the liver were elevated in plasma, with miR-122 and miR-192 showing higher levels after acetaminophen treatment (26).
In line with previous studies, our research showed that miR-122 was highly expressed in the precentral and periportal areas. Statistical analysis of miR-122 and miR-192 results in this study showed that 60% of miR-122 and 87.4% of miR-192 were overexpressed by dose and time (Figure 2). These results indicated that miR-122 changes are diagnosed earlier than hepatic standard markers.
Antoine et al. investigated the biomarkers to identify patients with the APAP-induced liver injury who were admitted for the first time to the hospital (24). They showed that the level of miR-122 as a highly specific liver biomarker increased in these patients. 17% of their patients developed liver injury while 83% did not, according to the serum ALT level (24). For miR-122, the median values for patients with liver injury were 3.69 (0.43 - 96.0) versus 0.21 (0.07 - 0.81) for the patients who did not (P¼ 0.002).
Because of their size, frequency, tissue specificity, and relative stability in plasma, miRNAs can be considered as available exclusive markers to monitor tissue damages. miR-122 as a liver-specific biomarker can be used to diagnose APAP-induced acute and chronic liver damage (13, 30).
The results of this study are consistent with those of previous studies that reported miR-122 and miR-192 changes are diagnosed earlier than hepatic standard markers. miR-122 is more sensitive than ALT in response to dose and time and the liver injury can be diagnosed in the early phases of the follow-up. Inherently, miRNAs are more easily detected than other proteins. As our study and others shown, circulating miRNAs are stable and can be extracted and examined in every serum sample. Moreover, considering that miRNAs remain longer in biological samples than proteins, they can be suitable diagnostic markers in the liver injury. Finally, miR-122 and miR-192 may be more specific for liver diseases in serum or plasma than ALT/AST enzymes.