1. Background
Epigenetic alterations are heritable modifications on the genome, which lead to changes in the gene function without affecting the nucleotide sequences. DNA methylation is the most common epigenetic change that plays a critical role in biological processes such as X chromosome inactivation, genomic imprinting, cell reprogramming during differentiation, and regulation of gene expression in mammalian cells. Considering its role in the regulation of gene expression, defects in DNA methylation may cause several diseases, including cancer (1-4).
It’s well-documented that DNA methylation patterns in tumor tissues differ largely based on their corresponding normal tissues. Cancer cells commonly represent a global hypomethylation in the whole genome along with regional hypermethylation (2, 3, 5). These hypermethylations usually occur at the promoter of genes encoding tumor suppressors, adhesion molecules, and DNA repair proteins in cancer cells (3, 6). Several studies argued that promoter hypermethylation via silencing of tumor suppressor genes (TSGs), DNA repair genes, and adhesion molecule genes (e.g. E-cadherin (CDH1 gene)) is correlated with tumor invasiveness and progress (5, 7, 8). Also, hypomethylation at the promoter region of oncogenes (e.g. urokinase plasminogen activator (uPA)) might cause their activation and tumor invasion (9, 10). As epigenetic alterations are reversible, today, there is a great interest in correcting these defects using epigenetic drugs. In this regard, DNA methyltransferases (DNMTs), which appear to be especially important for epigenetic changes, have become the focus of interest (4, 11-14).
Several chemical compounds have been identified as DNA demethylating agents, which generally act by inhibiting DNMTs. DNMT inhibitors, are divided into nucleoside and non-nucleoside analogues. Amongst nucleoside analogues, 5-azacytidine (Azacitidine), 5-aza-2’-deoxycytidine (decitabine), have been approved by the FDA for the treatment of myelodysplastic syndrome.
A number of clinical trials are performing on the efficacy of these drugs. Myelosuppression and neutropenic fever are reported as common side effects of these drugs (4, 14, 15). A number of non-nucleoside analogues are reported to have inhibitory effects on DNMTs, such as procaine, procainamide, hydralazine, RG108, and, recently, benzothiophene, with modest effects compared to nucleoside analogues: meanwhile, cytotoxicity is less common with these drugs.
Several in vitro studies have investigated the efficacy of these compounds on decreasing the promoter methylation and inducing the expression of TSGs in cancer cells (4, 14, 15). On the other hand, there are studies that have investigated the off-target effects of epigenetic drugs. Chik and Szyf (16) showed that treatment with decitabine can induce the expression of pro-metastatic genes (e.g. uPA) and increase the invasiveness of MCF-7 cancer cells. Many attempts have been made to find new DNMT inhibitors with more efficacy and lower toxicity. Discovering three-dimensional structures of DNMTs and the use of in silico structure-based screening approaches such as computational molecular docking and quantitative structure-activity relationship (QSAR) analyses provide the ability for high-throughput screening of many natural and synthetic compounds. In this manner, not only natural compounds are screened for DNMT inhibition, but also current FDA-approved medicines can be subjected to screening in a dug repurposing study (17, 18). Recently, olsalazine has been identified as an active DNMT inhibitor by in silico methods, and its capability in the hypomethylation of DNA has been demonstrated in the cellular context (19, 20). Olsalazine (Azo-disalicylate) is an approved drug in which two molecules of 5-aminosalicylate are linked by an azo bond and is used as an oral anti-inflammatory drug for the treatment of ulcerative colitis (21). Some studies have demonstrated that olsalazine can inhibit tumor growth and induce apoptosis in a rodent model of colorectal cancer (22). Although a previous study by Benno et al. (23) had shown that olsalazine may induce mitogenic actions in the rat intestinal epithelial cell. Hence, more evidence is needed.
2. Objectives
Therefore the present study was designed to investigate the cytotoxic effect of olsalazine on non-invasive breast cancer MCF-7 cell line and also its effect on the expression of CDH1 (an intercellular adhesion molecule gene) and uPA (an oncogene) genes, compared to decitabine.
3. Methods
3.1. Cell Line and Reagents
Human breast carcinoma epithelial-like cell line MCF-7 (NCBI# C135) was obtained from the Pasteur Institute of Iran (Tehran, Iran). Roswell Park Memorial Institute (RPMI-1640) medium, fetal bovine serum (FBS), penicillin-streptomycin solution, phosphate buffered saline (PBS), 0.05% trypsin-EDTA, and 0.2% trypan blue were purchased from Bioidea Company (Tehran, Iran) as sterile liquids. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell viability assay kit was obtained from Bioidea Company (Tehran, Iran). The 5-Aza-2’-deoxycytidine (decitabine) and 3,3’-Azobis 6-hydroxybenzoic acid (olsalazine) sodium powders were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Santa-Cruz Biotechnology (Dallas, TX, USA), respectively.
3.2. In Vitro Cell Viability Analysis by MTT Assay
Breast cancer cells were grown in RPMI-1640 medium (Bioidea Company, Iran) supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL Streptomycin. Cells were maintained at 37°C in a 95% humidified atmosphere containing 5% CO2.
The stock solution of decitabine was prepared in dimethylsulphoxide (DMSO)-milli-Q water solvent mixture (1:1), whereas olsalazine was dissolved in milli-Q water and kept at -20°C. Stock solutions were further diluted to working solutions in RPMI-1640 medium prior to use.
To determine IC50 values of drugs (the necessary concentration of a drug to achieve 50% inhibition in vitro), cells were seeded in 96-well microtiter plates at 5 × 103 cell/well density. After 24 h of incubation, cells were treated with decitabine or olsalazine at different concentrations (i.e. 0.1, 0.3, 1, and 3 mM) prepared in 100 µL RPMI-1640 medium supplemented with 10% FBS. No treatment was applied to control cells. Then, after 24 h of treatment, cell viability was assessed using a MTT assay kit (Bioidea Company, Iran) according to the kit’s protocol. The absorbance was measured at 570 nm by a microplate reader (BioRad, USA), and the results were expressed as the mean of three replicates. Cell viability was determined as the percentage of viable cells in treated samples relative to untreated control and plotted in a graph against drug concentration. The IC50 values were calculated accordingly and used to decide the optimum doses of the drugs for further investigations.
3.3. Relative Gene Expression Analysis by Real-Time PCR
To analyze the effect of drugs on CDH1 and uPA gene expression at the transcriptional level, MCF-7 cells were seeded in 24-well plates at 7.5 × 104 cell/well density. After 24 h of incubation, cells were treated with sub-lethal concentrations of olsalazine (1.5 mM) and decitabine (3 mM), prepared in 500 µL RPMI-1640 medium supplemented with 10% FBS. Untreated cells were considered as control. After 24 h, treatment cells were harvested. RNA extraction and DNase I treatment was performed using Quick-RNA MicroPrep kit (Zymo Research, USA), as per the manufacturer’s instructions. A total of 1200 ng RNA was reverse transcribed by the HIGH CAPACITY cDNA Reverse Transcription kit (Applied Biosystems, USA) according to the manufacturer’s instructions using a random hexamer.
The cDNA templates were subjected to a quantitative polymerase chain reaction (Q-PCR) to assess the expression of CDH1 and uPA transcripts. For Q-PCR, 2 µL of cDNA was used in a 20 µl reaction mix containing 1X Power SYBR Green PCR Master Mix (Life Technologies, USA) and 0.125 µM of specific primer pairs. The primer sequences used in this study are provided in Table 1. The reaction was performed in an ABI step one plus real-time PCR system (Applied Biosystems, USA) using the following conditions: a first denaturation step for 10 min at 95°C, followed by 45 repeats of the following cycle: 95°C for 15 sec, annealing at 53°C for 15 sec, and extension at 72°C for 20 sec. After the 45th cycle, an optional denaturation and renaturation step was carried out for 15 sec at 95°C and 1 min at 60°C, followed by a melt curve step ramping from 60°C to 85°C, rising 0.5°C per sec. The relative expression ratio of the target genes was computed based on their real-time PCR efficiencies (E) and the crossing point values (CP), using Pfaffle’s equation (24). Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) was used as the reference gene to normalize the data. No template control (NTC) was included in experiments. Each sample was analyzed in duplicate, and all experiments were carried out thrice independently.
| Primer Name | Primer Sequence | Product Length, bp |
|---|---|---|
| CDH1F113 | 5’-TCGCTTACACCATCCTCAGCCA-3’ | 113 |
| CDH1R113 | 5’-ACTCTCTCGGTCCAGCCCAGT-3’ | 113 |
| UPA104F | 5’-CCAAAGGCAGCAATGAACTT-3’ | 104 |
| UPA104R | 5’-GTTGCACCAGTGAATGTTGG-3’ | 104 |
| GAPDHF113 | 5’-CTCAACTACATGGTTTACA-3’ | 113 |
| GAPDHR113 | 5’-AAGATGGTGATGGGATTT-3’ | 113 |
List of Real Time PCR Primer Sequences
3.4. Statistical Analysis
The results are expressed as mean ± SEM (standard error of the mean). Data analysis was performed using Statistical Software for the Social Sciences (SPSS) version 16.0. One way ANOVA followed by Tukey post-hoc test and Pearson correlation analysis were used to analyze the significance between different values. Statistical difference was considered when P value < 0.05.
4. Results
4.1. Effect of Epigenetic Drug Candidates on MCF-7 Cell Viability
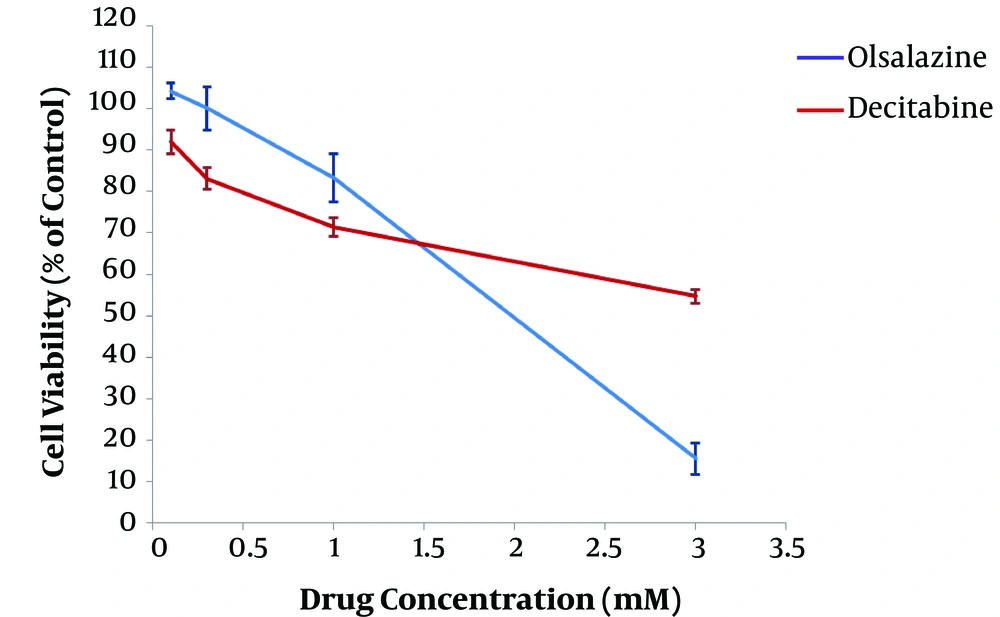
After 24 h of treatment, the effect of epigenetic modulator candidate (olsalazine), compared to the epigenetic drug (decitabine), on MCF-7 cell viability was assessed using standard colorimetric MTT assay. The results revealed an inverse association between cell viability percentage and concentration of drugs, indicating a dose-dependent effect (Figure 1). The IC50 values of olsalazine and decitabine were obtained about 1.75 mM and more than 3 mM in the MCF-7 cell line, respectively. The concentrations below the IC50 values, i.e., 1.5 mM and 3mM for olsalazine and decitabine, respectively, were considered for treatment in further experiments.
4.2. Effect of Epigenetic Drug Candidates on the Expression of CDH1 and uPA
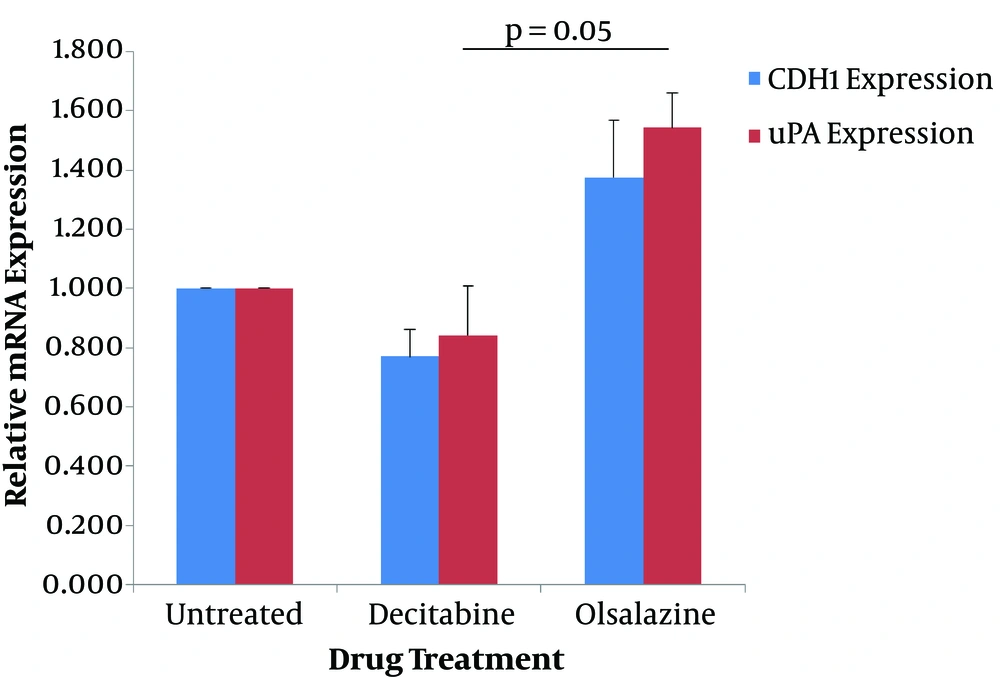
The effect of epigenetic modulator candidate (olsalazine) and the epigenetic drug (decitabine) on the expression of CDH1 and uPA genes was determined at the transcriptional level by quantitative real-time PCR method. For this reason, MCF-7 cells were treated with sub-lethal concentrations of olsalazine (1.5 mM) and decitabine (3 mM) for 24 h, and the extracted RNA was subjected to Q-RT-PCR. The results showed that olsalazine could increase the expression levels of CDH1 and uPA by 1.38-fold and 1.54-fold, respectively, compared to untreated control cells (Table 2). In contrast, treatment with decitabine could decrease the expression of CDH1 by 0.77-fold and of uPA by 0.84-fold compared to the untreated cells (Table 2). Although the results concerning three independent experiments were not significant, statistical analysis showed a significant difference between treatment with olsalazine and decitabine in uPA expression (Figure 2).
| Treatment | Relative Expression Levels | |
|---|---|---|
| CDH1 Expression | uPA Expression | |
| Untreated | 1.00 ± 0.00 | 1.00 ± 0.00 |
| Decitabine | 0.77 ± 0.09 | 0.84 ± 0.17 |
| Olsalazine | 1.38 ± 0.19 | 1.54 ± 0.12 |
Relative Expression Levels of CDH1 and uPA in MCF-7 Cells After Decitabine/Olsalazine Treatment Compared to Untreateda
Effect of decitabine and olsalazine on expression of CDH1 and uPA in MCF-7 cells. MCF-7 cells were treated for 24 h with olsalazine and decitabine, at 1.5 mM and 3 mM, respectively. The expression levels of CDH1 and uPA were determined by Q-RT-PCR. The relative mRNA expressions were represented as the mean of three separate experiments. Error bars represent the standard error of means. P < 0.05 was statistically considered as significant.
5. Discussion
Considering the important role of DNA methylation in the epigenetic regulation of gene expression and due to the reversible nature of epigenetic changes, several studies have focused on correcting these defects via inhibiting DNMTs (13). Recent studies mentioned olsalazine as a new potent candidate for DNMT inhibition with the potential to be used in cancer epigenetic therapy (19, 20). Therefore, the present study was designed to assess the cytotoxic effect of olsalzine on MCF-7 cells and also its effect on the expression of CDH1 and uPA (two cancer-related genes) compared to decitabine.
In the present study, human breast cancer cell line MCF-7 was used, as it is a non-invasive human breast carcinoma epithelial-like cell line in which the uPA gene is not expressed due to hypermethylation of its promoter region (10).
To determine the cytotoxic effect of both drugs on MCF-7 cells the standard MTT assay was used. The results of cytotoxicity assay showed that decitabine had no significant effect on MCF-7 cell growth at concentrations below 100 µM (data have not shown); Meanwhile, a dose-dependent effect was observed in higher concentrations. The IC50 value of decitabine was more than 3 mM in MCF-7 cells. Consistent with the results of the present study, Kastl et al. (25) showed that decitabine had no significant effect on MCF-7 and MDA-MB-231 cell growth in concentrations between 0.5 - 8 µM. In contrast to the findings of the present study, Ari et al. (26) have reported an IC50 value of 10 µM after 48 h of treatment of MCF-7 cells with decitabine, which is lower than the IC50 value of decitabine in the present study. Also, Kar et al. (27) have reported an IC50 value of 15 µM after 24 h of treatment of MCF-7 cells with decitabine.
Although cytotoxicity assay of olsalazine on MCF-7 cell showed no significant effect on MCF-7 cell viability at concentrations below 300 µM (data have not shown), a dose-dependent effect was observed in higher concentrations. The IC50 value of olsalazine was obtained about 1.75 mM in MCF-7 cells, suggesting that it might be more toxic than decitabine. Also, MendezLucio et al. (20) reported that olsalazine did not have a significant effect on iHO1 cervical cancer cells at concentrations of 0.1 and 10 µM. However, the findings of the present study are in contrast to their results, that the toxicity of olsalazine against iHO1 cells was lower than decitabine. A previous study by Benno et al. (23) suggested that olsalazine may induce mitogenic actions in rat intestinal epithelial cells. Although other studies have shown that olsalazine is able to inhibit cell growth and induce apoptosis in bovine endothelial cells and colorectal cancer cells (22, 28).
The present study intended to investigate the effect of olsalazine and decitabine on the expression of CDH1 and uPA genes by the Q-RT-PCR method. The results showed that treatment of MCF-7 cells with 3mM decitabine may decrease the CDH1 expression, although it was not statistically significant. Previous studies demonstrated that decitabine, as a DNMT inhibitor, can induce the expression of hypermethylated silenced genes by a dose and time-dependent manner (29, 30). Also, Kastl et al. (25) reported that the treatment of MCF-7 cells with 0.5 - 8 µM decitabine can increase the expression of GAPDH. This is an important finding because in our experiments, we used GAPDH as the endogenous control, and increasing its expression may affect the expression analysis.
The present study also showed that treatment of MCF-7 cells with 1.5 mM olsalazine resulted in a non-significant increase in CDH1 expression. In line with the findings of the present study, MendezLucio et al. (20) indicated that olsalazine at concentrations of 0.1 and 10 µM could induce the expression of methylated and silenced green fluorescent protein (GFP) gene in iHO1 cervical cancer cell. Meanwhile, in the present study, we did not find any significant difference between the effect of olsalazine and decitabine on CDH1 expression.
Concerning the effect of decitabine on the expression of uPA in MCF-7 cells, we found a non-significant decrease. In contrast, previous studies have shown that decitabine can induce the expression of hypermethylated genes (e.g. uPA in MCF-7 cell line), which in turn increases their invasiveness (16, 26). This difference can be attributed to the effect of decitabine on GAPDH expression, as discussed previously (25).
We also found that olsalazine may increase the expression level of uPA in MCF-7 cells and in this case, a significant difference was found between olsalazine and decitabine. As per previous studies, uPA is not expressed in MCF-7 cells because of its promoter hypermethylation (10). However, as reported by several studies, uPA gene expression may be induced by the use of DNMT inhibitors (16, 26, 31). This suggests that, despite the efficacy of epigenetic drugs in reverting aberrant epigenetic changes, their off-target effects should be considered in epigenetic therapy.
In summary, this study demonstrated that olsalazine was more cytotoxic than decitabine in MCF-7 cancer cells. Also, compared to decitabine, olsalazine could increase the expression of CDH1 and uPA genes, suggesting that olsalazine may have more ability to inhibit DNMTs than decitabine, although further studies are needed.