1. Background
Botulism is a potentially fatal disease caused by botulinum neurotoxin (BoNT), which is known as the most potent natural toxin, subsequently called “the poisonous toxin” (1-3). The family of BoNTs consists of seven districts serotypes (A-H). There are also newly found serotypes such as BoNT/H, which is believed to be a chimeric toxin (BoNT/FA). All these serotypes are similar in structure and function but are antigenically and immunologically different (4-8). There are also newly found serotypes such as BoNT/H, which is believed to be a chimeric toxin (BoNT/FA). All of these serotypes are similar in structure and function but are antigenically and immunologically different (4-8). Recent studies also represent another new serotype named BoNT/X, whose sequence is found to be the least identical to other serotypes (9). Despite the similarities in function, all of these serotypes are distinct from each other in size, target protein or site of action at the same protein, and potency and duration of action. Serotypes A, B, E, and F cause human botulism among which serotype A is the one which has been studied more than the others because of its higher potency and antigenicity (7, 10-12). BoNTs cause flaccid paralysis by inhibiting the release of acetylcholine neurotransmitter from the presynaptic terminals into the neuromuscular junctions (13-16). The synthesis of BoNTs, which mostly takes place in anaerobic Clostridium botulinum, primarily occurs as the preliminary formation of a single chain polypeptide with the molecular mass of 150 kDa, which post-translationally and with proteolytic cleavage changes into the active toxin that is a protein complex consisting of a 50-kD light chain and a 100 kDa heavy chain linked to each other by a disulfide bond and other non-covalent interactions (17, 18). The light chain is a zinc-dependent endopeptidase with catalytic activity, which prevents the neurotransmitter-filled vesicles from fusing to the plasma membrane. The heavy chain is divided into two domains, each of them located at one terminal of the chain. The one located at the C-terminal (50 kDa-HC) is responsible for binding to the receptor(s) of presynaptic membranes, and the N-terminal domain (49 kDa-HN) can form a channel across the endosomal membrane under acidic conditions to translocate the light chain into the cytosol (6, 13, 19-25).
The efforts to fight botulism and its severe damages throughout history, especially its use in bioterrorism, have led to the production of variable vaccines to provide enough protection against the toxin (26, 27). Though there has never been an FDA-licensed vaccine for botulism so far, there are examined suggestions to this plot such as the use of botulinum toxoids of serotypes A-E, which seems to be secure but under hesitancy due to the perilousness of production and preparation methods. Toxoids are chemically inactivated by formalin, which makes them painful for recipients. Despite the mentioned flaws, botulinum toxoids had been used for nearly 60 years for individuals who were at the risk of botulism (7, 8, 16, 28) and eventually, because of the above-mentioned flaws, this use came to a halt by the Centers for Disease Control and Prevention in 2011 (29). Another suggested form of vaccination against botulism is to use recombinant fragments of toxins to elicit immunogenic reactions in the recipient organism. The synthesis of the genetic sequence, which encodes a fragment of the toxin and transforming it into an expression system by the aid of molecular biology techniques can lead to the production of the protein, which is not also a recombinant form of the fragment with the same abilities, which brings almost no harm to the recipient due to the lack of hazardous properties of the original toxin. An example of this method is to isolate the encoding gene of the binding domain of the heavy chain, thereby achieving the production and expression of a translated heavy chain of recombinant botulinum neurotoxin serotype A (rBoNT/A-Hc), which lacks the translocating and catalytic activity of the toxin. As a result, this method turns out to be the least potent method of botulinum vaccination. After the binding of rBoNT/A-Hc to the receptor as an antigen, antibodies, which block the original toxin and subsequently raise the immunity level of the receiving organism, would be extracellularly produced. Simple production is another remarkable advantage to this method (7, 30, 31).
Among all expression systems, Pichia pastoris yeast is the desirable one due to its many advantages as a eukaryotic host, such as utilizing the powerful AOX1 promoter for protein expression, high-level expression, and convenient optimization of expression conditions (32). Different strains of Pichia pastoris have been evaluated as expression systems in different studies, among which x-33, GS115, and PichiaPink are of the most common ones used in recombinant protein studies and researches
2. Objectives
In order to prevent a severe syndrome such as Botulism, developing efficient vaccines against it is a necessity. Efforts have been made to accomplish this throughout time; however, some have discontinued due to the risks and unreliability of their production and usage
3. Methods
3.1. Gene Synthesis and Vector Construct
The DNA sequence of the binding domain in BoNT/A was isolated from the whole toxin sequence (code: EF470981) in FASTA format from NCBI and then synthesized by ShineGene Inc., China. The selected vector for the X-33 host strain was pPICZ A, and the one for PichiaPink strains was pPINK-HC. Both vectors were the products of Invitrogen Corporation. The synthetic gene was ligated into pPICZ A vector by T4 DNA ligase (Takara Co., Japan), in the nanobiotechnology research lab of Shahid Beheshti University, Tehran, Iran, where all the following methods and tests were done. The vector was treated by the KpnI restriction enzyme at the 5´ end and XhoI restriction enzyme at the 3´ end to develop compatible ends. Also, for ligating the same fragment into pPINK-Hc vector, the restriction enzymes SphI and KpnI were used at the 5´ and 3´ ends, respectively. The latter vector lacks the 6xHis-tag sequence, which is necessary for affinity chromatography, so the tag sequence was also added to the construct by PCR primers. Both ligated products were transformed to E. coli DH5α by heat shock at 42°C, and the resulted E. coli colonies were screened by colony PCR.
3.2. Transformation of P. Pastoris
Both recombinant vectors (extracted from PCR-confirmed colonies) were linearized with AflII restriction enzyme (Takara Co., Japan); afterward, each one was transformed to the desired strain of Pichia pastoris to be integrated into the genome of five Pichia pastoris strains; X-33, and four strains of PichiaPink kit. The transformation was performed according to the protocol of EasySelectTM Pichia Expression Kit Manual by Invitrogen Corporation, US. The transformants were spread in Yeast Extract Peptone Dextrose medium (YPD) plates and were incubated at 30°C. After 8-10 days, white colonies that had grown on the plates were streaked onto YPD plates. To screen the positive colonies, PCR was performed on the extracted DNA of the recombinant yeasts.
3.3. Screening of Expression Colonies
To choose the most efficiently expressing colony, all recombinant verified yeast colonies were screened by cultivating in micro-scale mediums before the macro-scale expression. After choosing the best expressing colonies, a loop-full of each one of them was primarily pre-cultured in 20ml of buffered glycerol complex (BMGY) medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate pH = 6, 1.34% YNB, 4 × 10-5% biotin, 1% glycerol) at 30°C in distinct shake flasks at the rate of 180 rpm for 24 hours. The BMGY volume of each shake flask was then increased to 100 mL. After another incubation for 24 hours, the grown cells were isolated from mediums by 15 minutes of centrifugation at 4°C, using a ROTOFIX32A rotor (Hettich universal 320 R, Germany) rotating at 8,000 RCF, in order to be transferred to the buffered methanol complex (BMMY) medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6, 1.34% Yeast Nitrogen Base (YNB), 4 × 10-5% biotin, 0.5% methanol) for induction. The grown cells of strains were resuspended in distinct shake flasks containing 100 mL of BMMY medium, which were afterward incubated under the same condition of the previous step. Methanol feeding for induction was done by the addition of 0.5% v/v methanol to the shake flasks per day. After 96 hours of incubation, cells were harvested by 15 minutes of centrifugation with the conditions mentioned earlier. Sampling was daily done to evaluate the amount of expression.
3.4. Cell Disruption
The harvested cells of each strain were dissolved in 100 mL lysis buffer (50mM sodium phosphate pH = 7.4, 1mM phenylmethylsulfonyl fluoride (PMSF) or other protease inhibitors for protecting the proteins from degradation, 1 mM Ethylenediaminetetraacetic acid (EDTA) and 5% glycerol) at 4°C. The prepared homogenate was injected into an ATS Pilot High-pressure homogenizer for cell disruption at 4°C under the pressure of 1,000 bar. After 8 cycles, it was centrifuged for 15 minutes at 10,000 rpm at 4°C. The resulted supernatant was collected and five-fold concentrated with wash buffer (Na2HPO4 20 mM, NaCl 300 mM, imidazole 20 mM pH = 7) using a tangential flow filtration (10 kDa Minimate TFF) from Pall Corporation, US.
3.5. Protein Purification
The protein purification steps were performed with an ÄKTAprime chromatography system, using a 5 mL the HisTrap column from GE Healthcare company, US, which traps the histidine-linked N-terminus of the intended protein. The column was washed with 10 column volumes of wash buffer to develop the necessary baseline. After injecting 500 µl of protein solution from the last step into the column, another 5 column volumes of wash buffer were applied to the column. Then a gradient mode was injected, using two distinct binding buffer solutions with different percentages of imidazole, to the column within 10 minutes to wash the intended protein from the column. The washed protein was obtained in 2.5 mL fractions.
3.6. SDS-PAGE and Western Blot Analysis
For performing SDS-PAGE a Mini-PROTEAN Tetra Cell from Bio-Rad, US, with 12% precast polyacrylamide gel was utilized. Necessary items, including load buffer and tank buffer, were prepared, according to the Bio-Rad instructions. The resulting protein bands appeared on the gel by Coomassie blue and silver nitrate staining. The densitometric analysis of the purified protein band on SDS-PAGE was performed by TotalLab Quant 12 software.
For analyzing the recombinant protein by Western blot, Bio-Rad blot module was used at 4°C for 2 hours to transfer the protein bands from SDS-PAGE to a polyvinylidene difluoride (PVDF) membrane )from GE Healthcare company, US( under the constant current of 30mA with transfer buffer (Tris base 25mM, glycine 192mM, methanol 20% v/v). After the transfer taking place, blocking of transferred protein bands was done by applying 3% Bovine serum albumin (BSA) to the membrane and incubating overnight at 4°C. This BSA solution also contained a 1:10000 dilution of Monovalent Botulinum Antitoxin Type A (ZABOT - A®) from Masoondarou Biopharma, Iran, dissolved in Phosphate Buffered Saline (PBS). The membrane was washed 3 times for 5 minutes each, with PBST (PBS with the addition of tween-20) and once with PBS. A mixed solution of 3% BSA and 1:1000 diluted Anti-horse IgG peroxidase conjugate (Sigma, A9262) in PBS was prepared in which the membrane was incubated for 2 hours at room temperature, and subsequently, it was washed with PBST and PBS just as it was performed two steps earlier. The bands on the membrane were visualized by 2 minutes of DAB (3,3'-diaminobenzidine tetrahydrochloride) substrate (Sigma Corporation, Japan) staining.
4. Results
4.1. Cloning
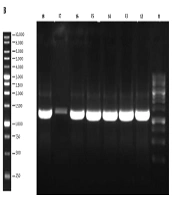
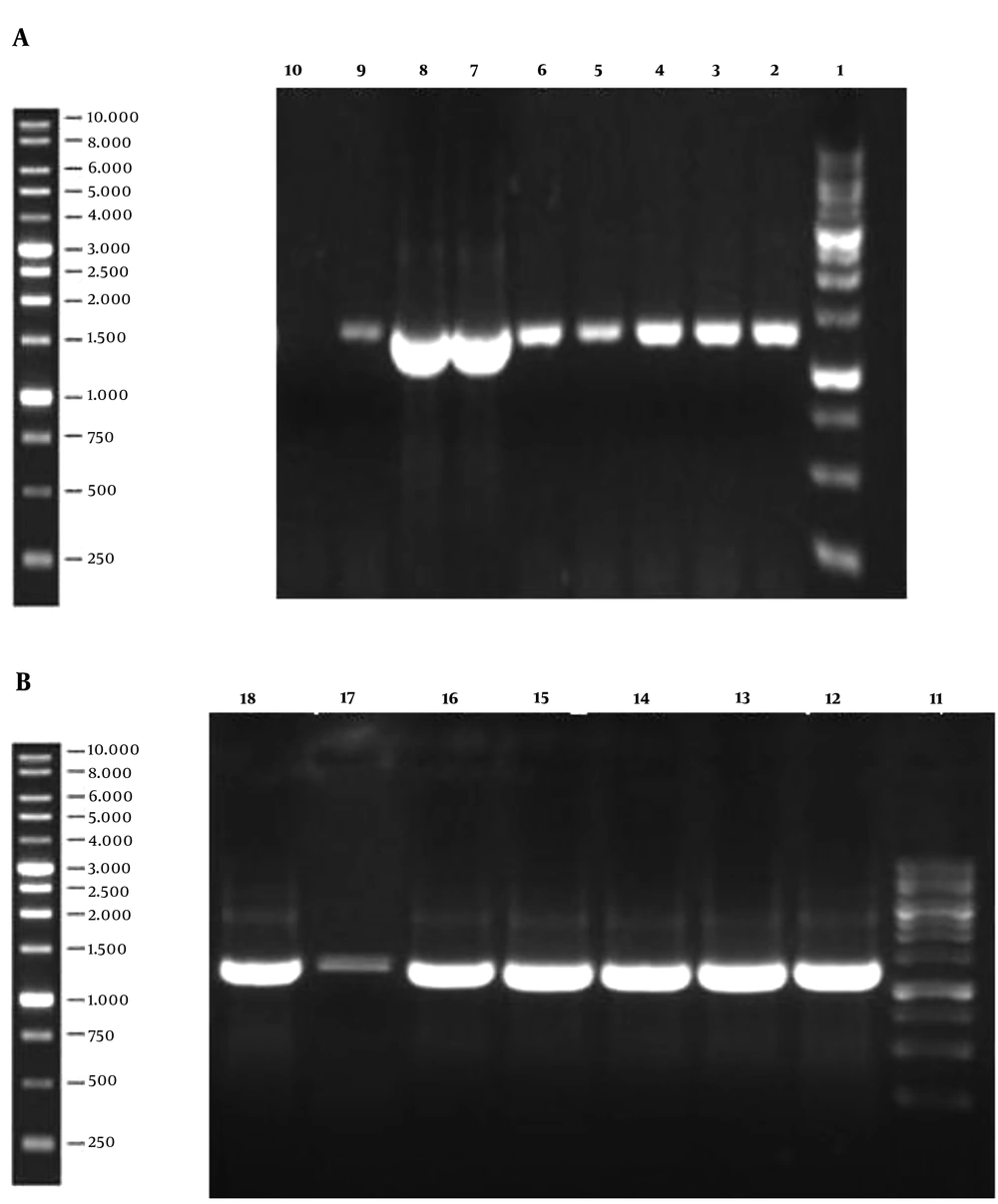
The 1,296 bp gene fragment of binding domain of BoNT/A was synthesized, which was afterward analyzed by GeneScript online software to optimize the cloning process. The gene construct inserted into the pPINK-Hc vector was designed to contain the His-tag sequence since the mentioned vector lacks this sequence. The modified gene fragment was cloned into both pPICZ A and pPINK-Hc vectors and it was subsequently transformed into E. coli DH5α. The resulted colonies were successfully confirmed to contain the recombinant vectors by loading the colony PCR products on agarose gel (Figure 1). After plasmid extraction of achieved colonies, pPICZ A and pPINK-Hc recombinant vectors were transformed into X-33 and four distinct strains of PichiaPink kit expression systems, respectively. After 8-10 days, there were white colonies on PichiaPink strains 2 and 4 and also X-33 YPD plates, which were the recombinant colonies since the color of the cells was changed from pink (the color of non-recombinant cells) to white. The YPD plates of PichiaPink strains 1 and 4 did not have any white colonies, and as a result, the recombination of these strains was not successful. PCR was performed on the genomic DNA extraction of transformants using specific primers, which approved the successful transformation of the recombinant vectors into the eukaryotic hosts (Figure 2).
Colony PCR Results of Recombinant pPICZ A (a) and pPINK-Hc (b) vectors on agarose gel; lane 1 and 11: ladder, Lanes 2-8, and 12-18: PCR result of recombinant colonies demonstrates successful cloning of the gene by the appearance of the 1,296 bp band on the gel. Lane 9: PCR positive control. Lane 10: PCR negative control.
The Result of PCR Performance on the Extracted Genomic DNA of Yeast Transformant. Lane 1: Ladder, Lanes 2 - 5: PichiaPink strain 2 recombinant colonies, Lanes 6 and 7: PichiaPink strain 4 recombinant colonies, Lane 8: X-33 recombinant colony, Lane 9: PCR positive control, Lane 10: PCR negative control.
4.2. Protein Expression
Protein expression took place in both micro and macro-scales. Micro-scale expression was done at low volumes (one-tenth of macro-scale volume) of mediums and less time to achieve an approximate expression level of each of the recombinant colonies. Furthermore, methanol induction in the micro-scale was more intense (four times more) in comparison to the macro-scale. Eventually, 3 out of 10 (2 of PichiaPink strain 2 and 1 of PichiaPink strain 4) colonies were selected as the best expressing ones, which were cultivated to express the recombinant protein in macro-scale. The pre-culture was prepared to generate enough biomass for cultivation in a glycerol-based medium. Methanol induction led to recombinant protein expression, which, based on western blot analysis performed; thereafter, resulted in intracellular expression of 46 kDa recombinant BoNT/A-Hc only by PichiaPink strain 2 and none of the other two strains. Daily sampling demonstrated that four days of methanol feeding would result in the optimum expression of recombinant protein, based on SDS-PAGE and western blot analysis. The western blot analysis result was indicated by the presence of the distinct reactive band of the recombinant protein (Figure 3).
Western Blot Analysis of Three Recombinant Yeast Colonies; Lane 1 and 3 are the product analysis of X-33 and PichiaPink strain 4 expression systems, respectively. The absence of 50 kDa band implies that the two strains have failed in expressing the recombinant protein. In contrast, the visible 50 kDa band in lane 2, which belongs to PichiaPink strain 2, indicates the successful recombinant protein production by this strain.
4.3. Purification of Recombinant Protein:
Affinity chromatography based on trapping the hexahistidine tag of the protein was the selected method to purify the expressed recombinant BoNT/A-Hc. At the first step, as the binding buffer was applied right after injecting the sample into the column, the contaminants were washed away, which appeared on the chromatogram as the first peak (Figure 4). The next resulting peak represents the target protein being eluted by a higher concentration of imidazole (500 mM), which competes with the his-tags for binding to the column. SDS-PAGE analysis was then performed for evaluating the purity of each of the achieved protein fractions. The results of SDS-PAGE demonstrated that one of the fractions contained only the purified recombinant protein, which appeared on the gel as a sole protein band with the correct molecular weight of 50 kDa (Figure 5). The result of densitometry analysis of this band indicated that the target protein was more than 95% pure and so the purification process was efficient. The final concentration of protein was determined 50 mg/L by Lowry protein assay (33).
SDS-PAGE Analysis of Protein Purification Obtained Fractions. Lane 1: 3 µg BSA. Lane 8 indicates the specific sole band of 46 kDa target recombinant protein. The other 6 lanes (2-7) represent other achieved fractions of protein purification process. As shown, all of these 6 fractions lack the sole band of recombinant BoNT/A-Hc protein.
5. Discussion
Developing vaccines against botulism disease is a serious concern, especially in countermeasure bioterrorism. The causative agent of botulism is botulinum toxin (BoNT), which is produced by Clostridium botulinum.
The main goal of this study was to isolate an immunogenic recombinant binding domain of BoNT serotype A (BoNT/A) by cloning it to an E. coli vector and then transferring the recombinant gene to the yeast Pichia pastoris to evaluate the expression of this protein by five different strains of Pichia pastoris for both accessing the best expressing strain and also achieving the expression of the verifiable recombinant protein. This protein is supposed to function as a vaccine against botulism.
Recombinant BoNT-Hc of other serotypes has also been produced (34, 35), but serotype A is the most studied one among distinct known serotypes of BoNT, and since it causes human botulism, it was chosen as the serotype of interest in this research. Pichia pastoris is the selected organism to express the product since it has a high potential for expressing the protein of interest in large quantities. There have been studies where E. coli was the expression system of BoNT-Hc, but some disadvantages, such as the possibility of the formation of inclusion bodies, were the reason for seeking more suitable expression systems such as Pichia pastoris (25, 31). Expressing this protein in Pichia pastoris, just as expressing it in E. coli comes with a limitation, which is the AT-riched sequence of BoNTs. This characteristic of the gene is incompatible with the optimum expression of clostridial genes in the expression systems mentioned earlier (36). Two strains of Pichia pastoris, including PichiaPink and X-33 have been evaluated as the proper expression systems to produce the recombinant binding domain due to the importance of choosing the right expression system, which depends on the biochemical and biological properties of the product (37). The PichiaPink strains have an advantage over X-33 strain as the desirable expression system. There are four PichiaPink strains that are distinct from each other in knockout proteases; PichiaPink strain 1 lacks the ade2 gene, and it is the parental strain of all the other three, which means all the other strains also lack the ade2. In addition to ade2, strain 2 is a pep4 knockout as well, which means it does not have the ability to synthesize proteinase A. The mentioned loss also leads to a decreased activity of proteinase B and a lack of carboxypeptidase Y activity. Strain 3 is a prb1 knockout, which leads to the prevention of proteinase B production. Strains 4 is a double knockout of pep4 and prb1, which makes it unable to synthesize both proteinase A and B. The last strain thus has the lowest protease activity among all four strains. Consequently, all described strains would reduce the degradations of expressed protein by proteases and would also lessen the need for protease inhibitors to save the product from degradation (38). There have been attempts to produce the recombinant BoNT/A-Hc in the GS115 strain of Pichia pastoris (25, 26, 35, 39), but due to the advantages of the PichiaPink kit mentioned above, the expression of the recombinant product has been evaluated in the four strains of PichiaPink. Also, despite the failure of expressing the recombinant BoNT/A-Hc by X-33, this strain is proved to be efficient in expressing the BoNT/Hc of other serotypes such as C (34).
In conclusion, the selected gene of BoNT/A-Hc was synthesized, optimized, cloned into E. coli vectors, transformed to Pichia pastoris strains, and after being expressed, it was purified and finally, analyzed by analytical techniques. It has also been demonstrated that the PichiaPink strain 2 is more desirable to be used as an expression system for this recombinant protein rather than X-33 and the other three strains of PichiaPink. The comparison between the yields of the products of the strains also does not show any desirable results in the other three PichiaPink strains and X-33 expression system, which declares the PichiaPink strain 2 as a more efficient one for this purpose.