1. Background
Biosurfactants, produced by a wide spectrum of microorganisms, are amphipathic molecules that reduce surface tension (1, 2). Biosurfactants are classified in lipopeptide, lipoproteins, glycolipids, phospholipids, and polymeric, according to their origin and nature of chemical structures (3, 4). Compared to chemo-synthetic surfactants, biosurfactants have been shown to have many advantages. In addition to being environmentally friendly, biodegradable, less toxic, non-hazardous, and have stability in extreme pH, salinity, and temperature (5). Hence, potentially can be used as surfactants and emulsifiers in the pharmaceutical (drug delivery), cosmetics (foaming agent), and food industries (emulsifier and stabilizer) (6, 7). Nowadays, despite the complicacy of producing biosurfactants, they are used in many cases other than chemical surfactants. Despite all these advantages, the greatest limitation of using biosurfactants on a large industrial scale is high production costs (8). Bacteria strain such as Bacillus, that can survive in harsh environments for a long time, are generally known as biosurfactants producers (9). Several studies have investigated the isolation of Bacillus species, as biosurfactant producers, from saline and hypersaline soil sample (10, 11).
Evaluating factors that contribute to biosurfactant production is the most important method to maximize biosurfactant production. Biosurfactants can be produced by different substrates such as oils, distillery and dairy wastes, and agro-industrial byproducts, which are economical (12).
2. Objectives
The current study aimed to isolate and identify biosurfactant producing bacterial strain from saline and hypersaline soil samples and to evaluate factors that contribute to biosurfactant production.
3. Methods
3.1. Screening for Biosurfactant Producing Bacterial Strain
Soil samples were collected from different locations of saline and hypersaline environments in Kerman, Iran. One gram of each soil sample was then added to 20 mL sterile normal saline and mixed well. Thereafter, 500 µL of each obtained sample was spread on a blood agar plate. The culture plates were then incubated at 30°C, and the colonies with the highest hemolytic halos were regarded as potential isolates for biosurfactant production. In the next step, the desired isolates were further purified by sub-culturing on nutrient agar medium to obtain axenic culture. The achieved pure isolates were preserved using glycerol 20% (v/v) at −80°C (12).
3.2. Evaluation of the Selected Isolate for Biosurfactant Production
3.2.1. Hemolytic Activity as a Preliminary Screening
The blood agar plates containing 5% v/v blood were used to seed the desired isolate, which was incubated at 37°C for 48 hours. Then, the plates were examined to form the clear zone around bacteria colonies, and the size of the clear zone was measured (13).
3.3. Biosurfactant Profile
At first, the selected isolate was cultivated in nutrient broth (NB) medium and incubated at 37°C for 96 hours. Then, samples were taken intervallic and analyzed for OD600 by photospectroscopy (UV-1800, Shimadzu CO, USA) followed by centrifugation (5000 rpm for 10 minutes) and determination of EI%, Oil spreading test, surface tension, and pH alteration (5).
3.3.1. Emulsification Measurement
Mineral oil (5 mL was added to equal valium of supernatant and vortexing at high speed (300 rpm) for 2 minutes. The emulsion stability was determined after 24 hours. EI% was determined using the following equation (5):
(Height: mm)
3.3.2. Oil Spreading Test
A volume of 100 µL of the supernatant (as mentioned above) was dropped onto the surface of the mineral oil. The diameter of the clear zone on the oil surface was measured and compared with 100 µL of un-inoculated culture media as the negative control (14).
3.3.3. Surface Activity Measurement
Tensiometer (KRUSS-K100, Germany) was applied to measure the surface tension of the culture supernatant. The values reported are the mean of three measurements (12).
3.4. Identification of the Isolated Bacterial Strain
The morphological characterization of the selected isolate was evaluated by macroscopic and microscopic evaluation. Also, using several biochemical tests, the strain of bacteria was further characterized (5, 15). For further identification of the isolate, analysis of the 16S rDNA gene was performed after extraction of genomic DNA by biomass heating. The obtained template DNA was then applied to prepare a PCR mix containing the primer pair of 27F (5'-AGAG TTTGATCCTGGCTCAG-3') and 1525R (5'-AAGGAGGTGATCCAGCC-3'). The Primus 96 advanced thermal cycler (PEQLAB, Erlangen, Germany) was employed for target gene amplification using the program of (a) 94°C for 3 minutes; (b) 30 cycles of denaturation at 94°C for 1 minute, 60°C for 45 seconds and 72°C for 90 seconds; and (c) the final extension at 72°C for 90 seconds. The desired samples containing the amplified 16S rDNA gene were sent to Takapouzist Co. (Tehran, Iran) for sequencing. After the determination of forward and reverse sequences, the complete sequence of the 16S rDNA gene was built using the software of GenRunner (version 3.01) and comprised other recorded genes in the GenBank of NCBI using BLAST software. The phylogenic tree of the selected isolate was drawn. The obtained sequences were then submitted to the GenBank of NCBI to get accession number (5).
3.5. Evaluating the Effect of Factors on Biosurfactant Production
To evaluate the medium components contributing to biosurfactant production, the selected strain was grown on nutrient broth with different vegetable oil carbon sources (almond oil, olive oil, castor oil, sweet almond oil, and hazelnut oil), nitrogen sources [yeast extract (1%), peptone, tryptone (1%), urea, ammonium nitrate (1%), and ammonium chloride (1%)], and cations [Na+ (0.1 w/v), Ca++ (0.1 w/v), Fe++ (0.1 w/v), Mg++ (0.1 w/v) and K+ (0.1 w/v)]. Withdrawn samples were analyzed every 12 hours to the surface activity measurement (5, 12, 16).
4. Results and Discussion
4.1. Hemolytic Activity
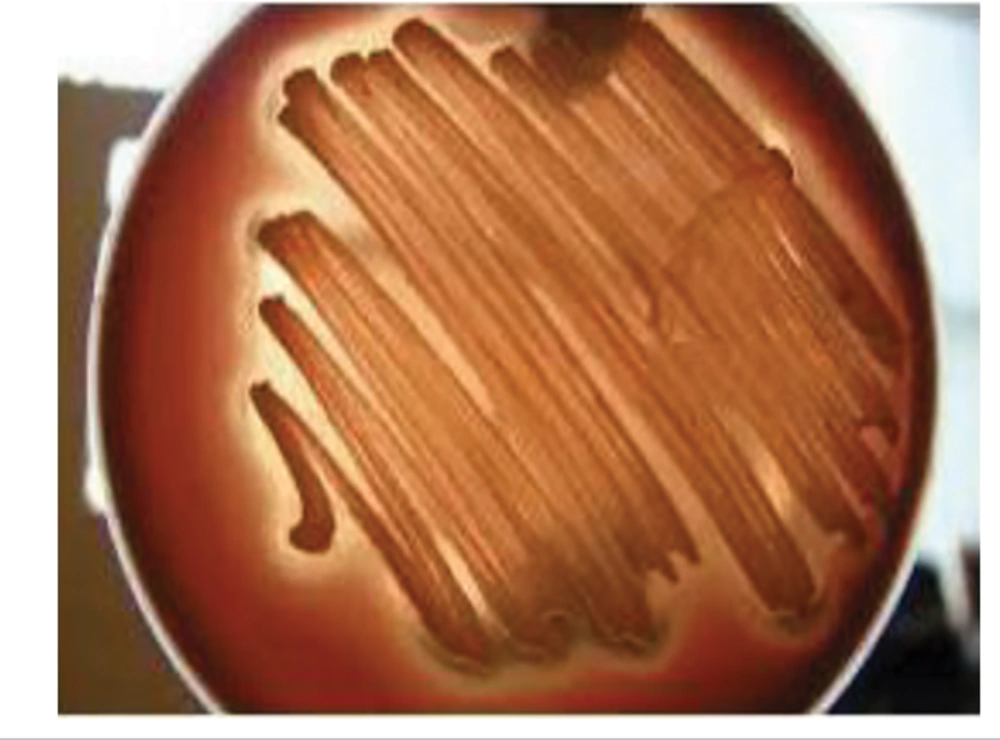
For all six isolated bacterial colonies (obtained from 10 collected samples), the hemolytic activity of the biosurfactant-producing bacterial (designated as M2) was verified on the blood agar medium by the production of halo around the related colony (Figure 1). Hemolytic activity measurement is applied as a primary method to screen biosurfactant production (17). The same process was done by Abouseoud (18) for biosurfactant activity of Pseudomonas fluorescens. The diameter of the hemolytic zone was used as an indicator.
4.2. Emulsification Index
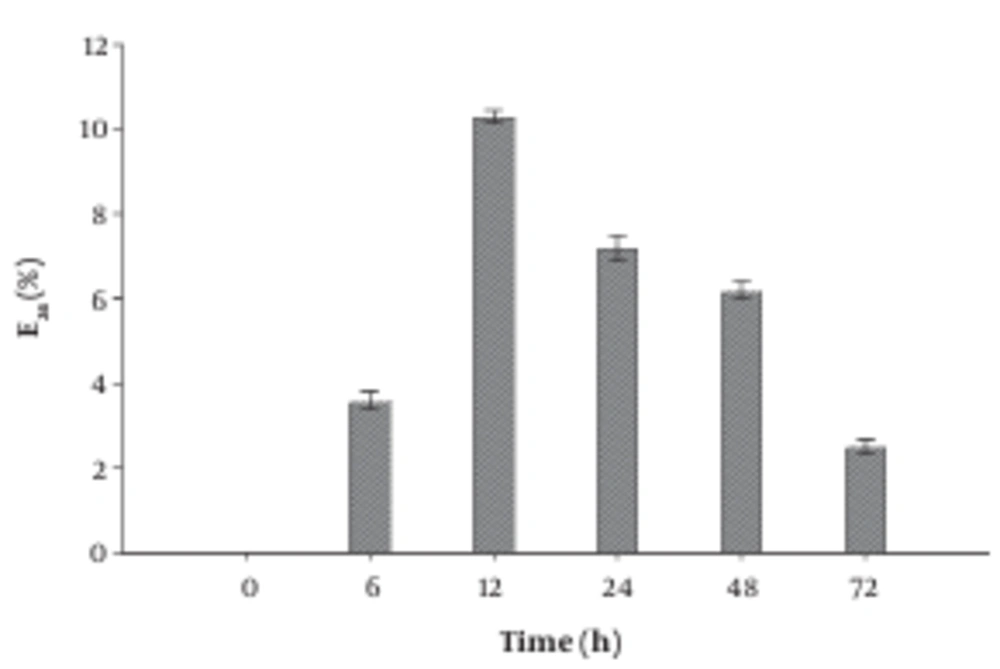
After cultivating the bacterial isolate for 72 hours, the produced supernatant was harvested at different time intervals and tested for the E24 index. As shown in the profile of E24 determination (Figure 2) the maximum E24 index recorded after 12 hours of cultivation, where the E24 index reached 10%. Similar results are reported by Cooper et al. (19).
4.3. Oil Spreading Assay
Oil displacement tests were also performed, which exhibited a clear diameter of 40 mm. Similar results are observed Sharma et al. (20), who isolated P. aeruginosa DSVP20, as biosurfactant-producing bacterial from petroleum-contaminated soil samples.
4.4. Surface Tension Determination
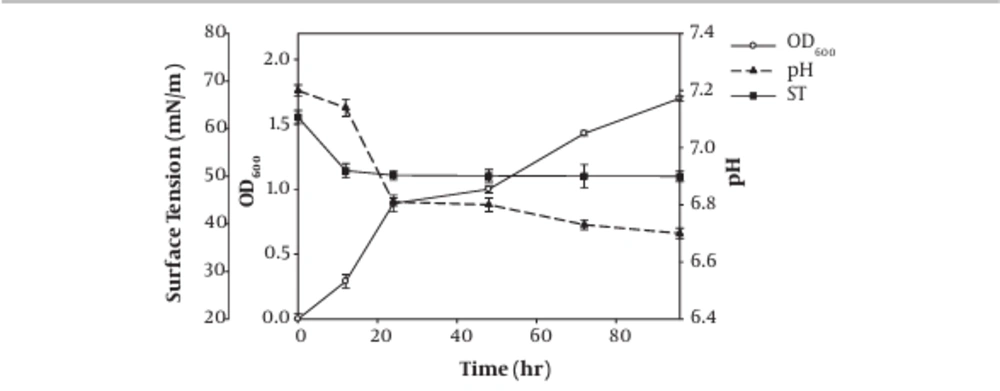
The results regarding surface tension measurement, as well as OD600 nm recording and pH estimation of the obtained supernatant of the isolate M2 during 96 hours incubation, are shown in Figure 3. The surface tension was dropped to 51.17 mN/m after 12 hours of incubation in a time-dependent manner. The effectiveness of the surfactant was determined by its ability to reduce the surface, and interfacial tension, so more ability in surface tension reduction was associated with more activity as an amphiphilic molecule (5). In the case of biosurfactant production from marine Streptomyces species B3, the surface tension was reduced during the five days after incubation and reached 28 mN/m (21). In our study, the surface tension decreased rapidly during the first day to 50 mN/m, so it can be argued that the strain can produce biosurfactant too rapidly, but the probably produced biosurfactant has low amphiphilic properties. Data on surface activity and emulsification index measurements (section 4.2) showed a correlation between emulsification index and surface activity, while when the minimal surface tension was achieved, the emulsification index reached its maximum (5).
4.5. Identification of the Selected Isolate M2
Morphological characteristics and biochemical tests of the isolate M2 were determined using standard protocols. It was a non-motile, Gram-positive, rod-shaped bacterium which formed a white to pale yellow colony on nutrient agar medium. The mentioned isolate represented positive reactions for catalase and oxidase and produced acid from maltose, sucrose, lactose, mannitol, glycerol, galactose, fructose, and glucose. This isolate hydrolyzed casein but couldn't hydrolyze gelatin and starch. Isolate M2 grew well at all concentrations of salt, which were tested (2.5 - 15% NaCl) and temperatures up to 40°C. However, the growth decreased at 50°C. The BLAST results of the 16S rDNA amplified gene revealed that isolate M2 was closely related to Bacillus atrophaeus with a similarity of 99%. The phylogenic tree of the obtained results is illustrated in Figure 4. The gene sequence was deposited in GenBank under the accession number of KF682367.
4.6. Effect of Factors on Biosurfactant Production
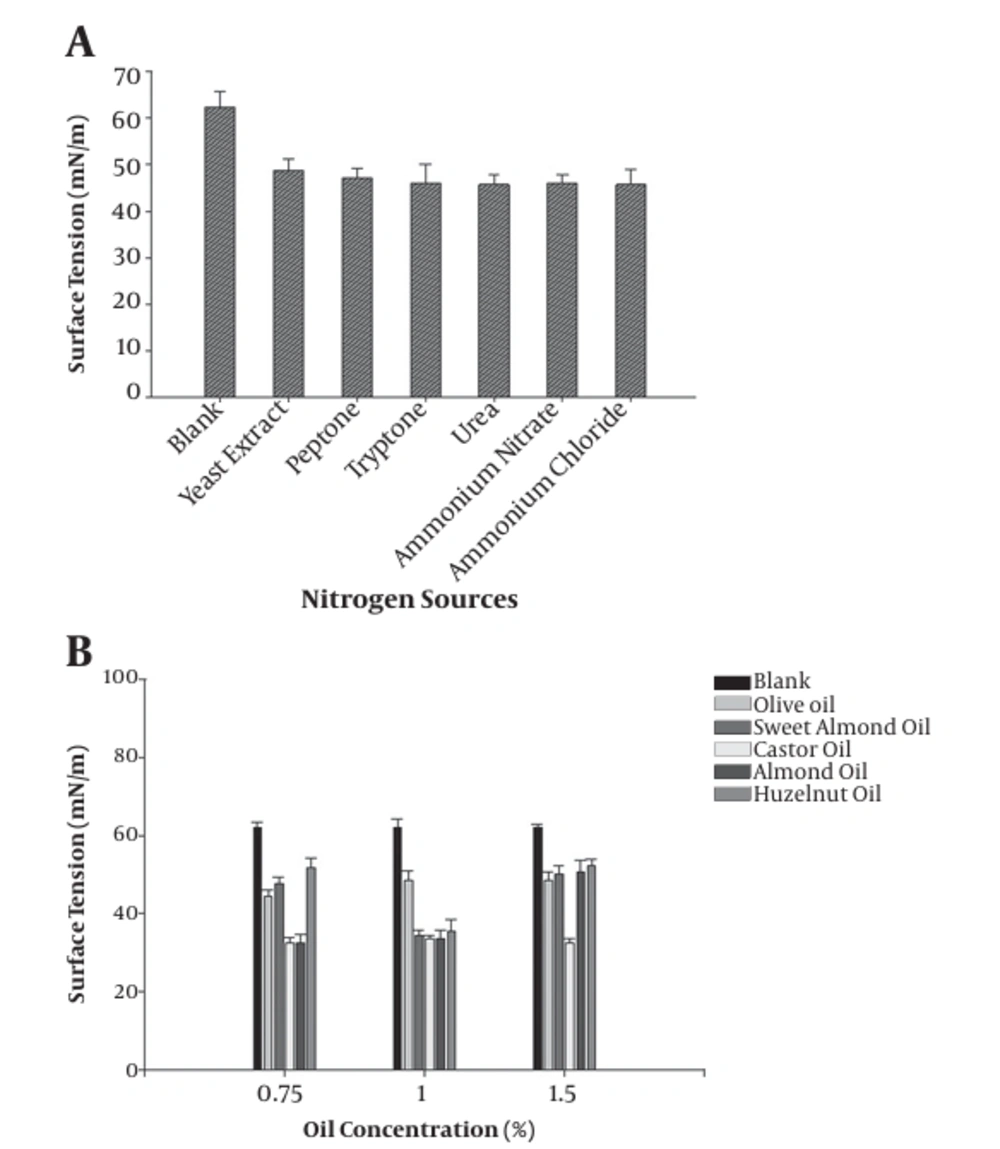
The effect of the addition of nitrogen source on the surface tension of the culture broth of B.atrophaeus is shown in Figure 5. All applied nitrogen sources reduced the surface tension compared to the un-inoculated culture medium. Also, the yield of biosurfactant was improved after adding vegetable oil, such as castor oil (1%) and almond oil (1%), to the culture medium (nutrient broth) (Figure 5). It's well-documented that the type and concentration of the carbon substrates, such as vegetable oils, can significantly affect the production yield of most biosurfactants (22). Dehghan- Noudeh et al. (23) reported that vegetable oils, like almond oil, induced biosurfactant production by Acinetobacter calcoaceticus more than other carbon sources. In another study, Khopade et al. (21) showed that a change in medium carbon source could change the amount of produced biomass and also biosurfactant production. In addition to carbon sources, nitrogen sources are other critical factors that regulate biosurfactant production (22). The obtained results of surface tension in the presence of each cation showed that the surface tension decreased (46 mN/m) in the presence of Na+ (0.1%) and Mg2+ (0.1%) compared to that of the blank (65 mN/m), respectively. Previously conducted studies reported that the highest biosurfactant production yield by P. aeruginosa was obtained when sodium nitrate was applied as the sole nitrogen source (22, 24).
5. Conclusion
The bacterial strain, B. atrophaeus, was successfully isolated from different locations of saline and hypersaline environments in Iran. B. atrophaeus could produce biosurfactants. Based on the results, nitrogen, carbon (castor oil, almond oil), and cation sources (Na+, Mg2+) are the main factors contributing to biosurfactant production by B. atrophaeus. Such biosurfactant producers would have great potentials in bioremediation activities and microbial enhanced oil recovery.