1. Context
1.1. Introduction to Drug Discovery
Drug discovery is a multidisciplinary process encompassing medicine, biotechnology, and pharmacology, during which new candidate drugs are identified. While several research teams work together and put in many efforts, most developed drugs do not acquire clinical approval because of either low efficiency or high toxicity in clinical studies (1). One of the most significant drug discovery issues is the absence of a way to directly monitor drugs’ targets and off-targets within cells and tissues. One reason for the failure of newly discovered drugs in clinical trials is that the drug’s targets are monitored via indirect methods such as studying downstream signaling pathways and cellular responses (2-4).
To acquire the approval of the US Food and Drug Administration (FDA), a drug must pass its development processes, including scaled-up and chemical analyses, pharmaceutical quality assessments, and finally, in vivo testing (5). Candidate drugs are evaluated for absorption, distribution, metabolism, and excretion (ADME), as well as for toxic properties in animals. Those candidates that pass these hurdles are formulated and assessed in clinical trials. Success in these trials will determine whether or not a drug receives FDA approval (5). However, costly procedures and infrastructure limit the testing scope of newly discovered chemical compounds with therapeutic potentials (6). Each year, a few chemicals are approved by the FDA, among them, even fewer are fully aware of their mechanisms of action (5). Indeed, the failure in acquiring clinical approval of a drug is a failure of decades of research and billion dollars of investment. In this regard, there is a crucial need to develop new methods that can increase drugs’ effectiveness in the human body. Drugs’ therapeutic effects in the body need specific interactions between the drug and a variety of biological macromolecules, exclusively proteins and nucleic acids. Consequently, recent advances in molecular biology and functional genomics ensued astonishing developments in this field, especially the emergence of effective and promising drugs.
1.2. Shortcomings of Drug Discovery Methods
Knowing the chemistry of a compound is necessary but not sufficient for developing an effective drug, and inadequate knowledge of the chemical properties of drugs, especially in biological systems, is a serious limitation in this area (7). There is no direct way to monitor the physical binding of a drug to a target in living cells (8-10). However, some techniques are available to ascertain physical interactions in cell lysates, including the protein oxidation protection method (11) and drug affinity and target stability assessment (12), which both are based on the protection of drugs via their interactions. Despite the identification of the drug targets of orphan ligands in cell suspensions (11), the application of these methods must be shown in monitoring dynamic intracellular events. In the presence of a selective and high-affinity ligand, in situ competitive testing can be performed to detect associated proteins or extracellular receptors and monitor target engagement (13).
2. Evidence Acquisition
2.1. The Importance of Proteomics in Drug Discovery
Considering the development of the genomics protocols containing instructions for making proteins, why is it necessary to examine proteomics? What is the need to study proteins directly?
The most significant answers to these questions are:
- DNA sequence information only provides a fixed and precise image of a variety of biochemical paths, while cellular life is a variable and dynamic process. A genome remains mostly intact while the cell’s proteins alter by turning genes off and on in response to environmental conditions. Finally, most gene products have unknown functions (14).
- Proteins are modified after transcription, which is necessary for them to play their roles, and gene sequences cannot describe these changes. On the other hand, the abundance of a protein in a cell does not indicate its level of activity because its amount often depends on post-translational changes (14).
- Many proteins are not always proportional to a unique mRNA (e.g., due to post-transcriptional changes, only parts of a specific mRNA may be translated to proteins, and for some of them, the translation process will not happen at all (14).
- The sensitivity of available devices only allows the detection of highly abundant mRNAs. However, in most cases, low-abundance mRNAs encode critically important regulatory proteins (14).
- Many crucial biological samples (such as urine, cerebrospinal fluid, and plasma) do not contain mRNAs. Moreover, in some specimens, mRNAs are decomposed and lost due to a prolonged interruption between the sampling time and mRNA testing (14).
Therefore, although the genome has a steady nature within cells or organisms, the proteome has variable nature according to intracellular and extracellular microenvironments. The characterization of all cellular proteomic states is called the proteotype, which is a channel between cellular genotype and phenotype. Accordingly, the prototype of a cell or tissue can influence drugs’ therapeutic efficiency (15). More than 10,000 gene products are expressed in variable degrees in various cells. They occur in diverse isoforms or various PTMs at different subcellular localizations. Nonetheless, because of the inherent complexity of the proteome, accurately defining the prototype seems to be difficult (16). In current proteomics studies, prototypes are described as lists of proteins and their semi-quantitative expressions (17). Further, a number of methods are available to assess some but not all post-translational modifications in cells (18, 19). The studies analyzing the functional mechanisms of bioactive compounds, like small-molecule drugs or peptides, aim to identify their possible binding partners in cellular extracts with a focus on affinity-based enrichment strategies (20-22). These methods can be mingled to detect various signaling biomarkers and pathways in different cells (23, 24).
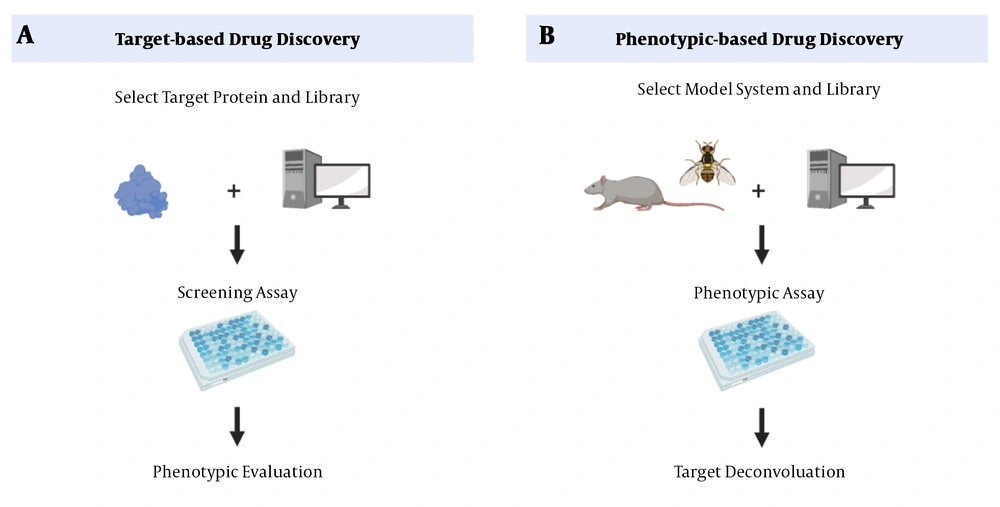
The main drug exploratory methods are target-based drug discovery (TDD) and phenotypic-based drug discovery (PDD) (Figure 1). In TDD, a target-based approach, the starting point is a defined molecular target hypothesized to have an essential role in disease pathogenesis, followed by a biochemical method to detect small molecules. The mechanism of action must still confirm candidates’ cellular activities and predict their possible side effects (25). A disadvantage of this approach is that proving the involvement of a protein in a particular biological pathway or a disease is time-consuming (25). On the other hand, PDD, a phenotypic-based approach, begins with examining a system’s phenotype. During this process, the small molecules that can trigger this phenotype are identified (3). However, the drug’s action mechanism, which is an essential criterion for drug development, is not determined (26). Although TDD has been a dominant approach in the pharmaceutical industry, there has been a revival of interest in PDD due to “omics” advances in recent years (27).
Drug discovery methods. A, target-based approaches (reverse chemical genetics). This method starts with identifying changes in molecular targets. B, phenotype-based approaches (forward chemical genetics). In PDD, after identifying the phenotype of a system, the small molecules that can stimulate the phenotype are identified (28).
Phenotypic screens expose candidate compounds to proteins in more biologically relevant contexts than screens involving purified proteins.
Since these screens measure cellular function without imposing preconceived notions of the relevant targets and signaling pathways, they offer the possibility of discovering new therapeutic targets. So, by determining specific targets and signaling pathways, they can discover new therapeutic targets (28).
The PDD approach has some challenges, the most important of which is the deconvolution of drugs’ mechanisms of action. So, several new methods have been extensively studied for identifying the budding target (28, 29). These strategies are based on mass spectrometry and expose small changes in the target’s stability following the binding of the compound (30, 31). The most important proteomic techniques include drug affinity responsive target stability (DARTS) (11), stability of proteins from rates of oxidation (SPROX) (32, 33), cellular thermal shift assay (CETSA), and TPP (34-36).
2.2. Drug Affinity Responsive Target Stability (DARTS)
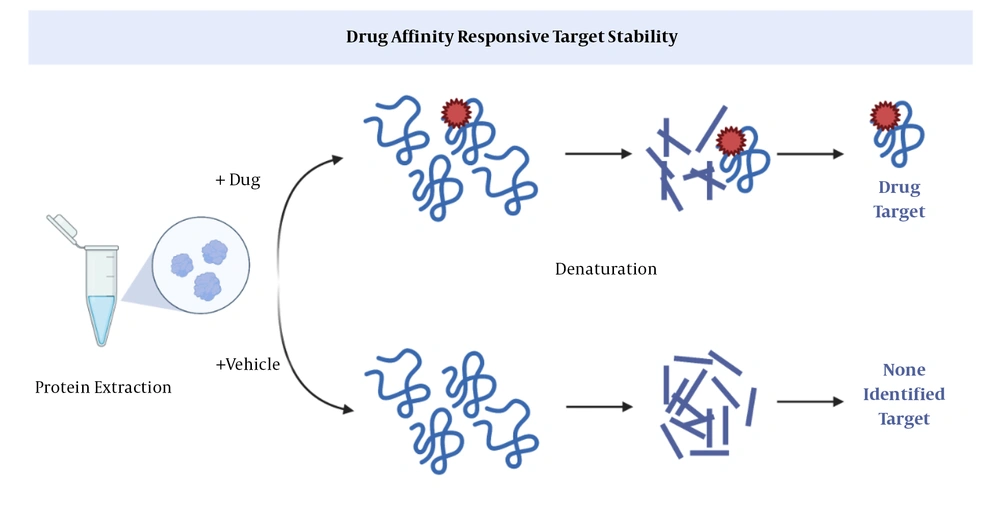
This method is based on limited proteolysis (37); the basis of which is the use of specific proteases that cleave a particular peptide bond in a protein (Figure 2). Via DARTS, it can be proved that the part of a protein exposed to a protease is protected by binding to a ligand. In general, the purpose of ligand binding is to stabilize a part or the whole structure of the target protein. A big drawback; however, is that changes in downstream proteins cannot be explored in the protein extract (11).
Drug affinity responsive target stability (DARTS) (11). After binding of a drug to a protein, proteases cannot cleavage the peptide, so the protein remains intact.
2.3. Stability of Proteins from Rates of Oxidation (SPROX)
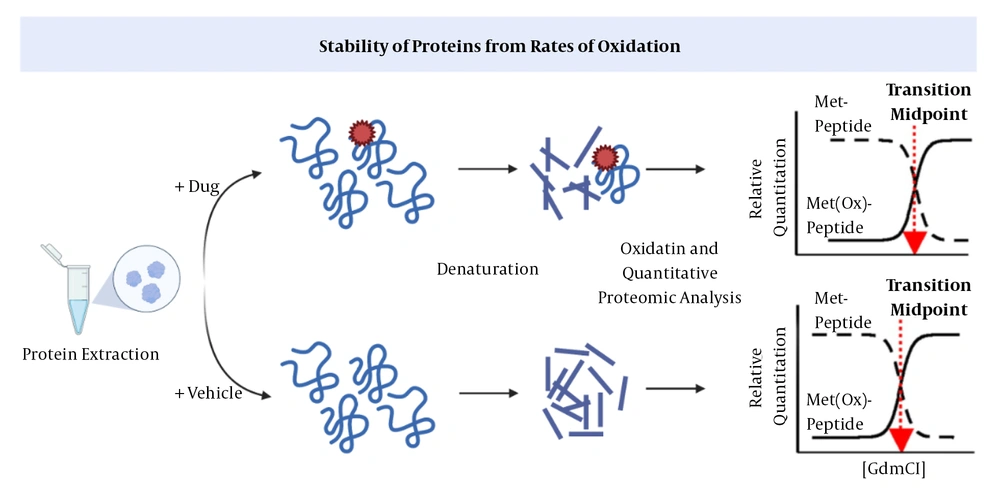
In SPROX, the aliquots of the protein are exposed to increasing concentrations of a chemical denaturing agent and then to methionines to determine the levels of oxidized and unfolded proteins (12, 32, 33). Following the binding of a ligand, proteins will be stabilized against chemical denaturation. This method, similar to DARTS, cannot identify the downstream proteins recruited by a drug (Figure 3) (12).
Stability of proteins from rates of oxidation (SPROX) (38). The drug-target interaction increases the protein’s stability against chemical oxidation.
2.4. Cellular Thermal Shift Assay (CETSA)
Researchers have established that monitoring differences in the thermal stability of proteins can be applicable for studying the ligand-binding process (39, 40). Regarding the absence of a method for cell-wide assessments and taking into account the recent advancements in measuring intracellular temperature changes (i.e., the CETSA method), researchers can evaluate target-ligand interactions in living cells (41, 42). As stated, CETSA allows studying target engagement in vivo. One of the advantages of using living cells is the possibility for monitoring pro-drugs’ mechanisms of import and activation (43).
The CETSA technique provides the opportunity to evaluate drug-protein interactions in the physiological environment, which is a breakthrough development and an essential step in drug discovery. Living cells are initially treated with the drug-protein complex, and then the target protein is identified based on changes in thermal stability (44, 45). The stability of a drug-target interaction is determined by the thermodynamic stabilization principle (i.e., excess energy is needed to separate the ligand after its binding to the protein) (34, 46-48).
In addition, the CETSA method can help to achieve important information on the potential targets of a wide range of different pharmacological agents. This approach was confirmed via several model systems to address problems such as drug activation, off-target effects, drug delivery, drug efficacy, and drug resistance in mammalian cells, as well as drug kinetics in animals. Also, the preclinical applications of developed drugs can be monitored in humans using the CETSA method. For example, CETSA can be used to determine the appropriate dose of a drug and the rate of drug resistance in cancer therapy. The adaptability of CETSA has delivered the method as a beneficial tool for drug research and development (41).
The CETSA process begins with treating cells with a drug or placebo, followed by heating the cells to unfold and precipitate the target protein. Then cell debris and aggregates are removed by centrifugation. Finally, using target-specific antibodies, the target protein is detected based on its thermal stability. The crucial steps of the CETSA method include heating (which triggers the denaturation and precipitation of proteins, except the ligand-bound proteins that are stabilized) and detection (in which stabilized proteins are distinguished from other proteins that are already denatured and precipitated during the heating process) (49). The most significant limitation of CETSA is that at the final step, stable proteins are identified by an antibody-based method (i.e. western blotting). Although western blotting facilitates the verification of predetermined proteins, it cannot detect non-specifically bound proteins (34). Therefore, since the CETSA is a limited antibody-based method, only a small number of specific proteins can simultaneously be studied, which is a significant disadvantage. A new approach has been developed by combining the CETSA method and mass spectrometry (MS). This new mass spectrometry-based technique is called TPP and could overcome this limitation.
2.5. Thermal Proteome Profiling (TPP)
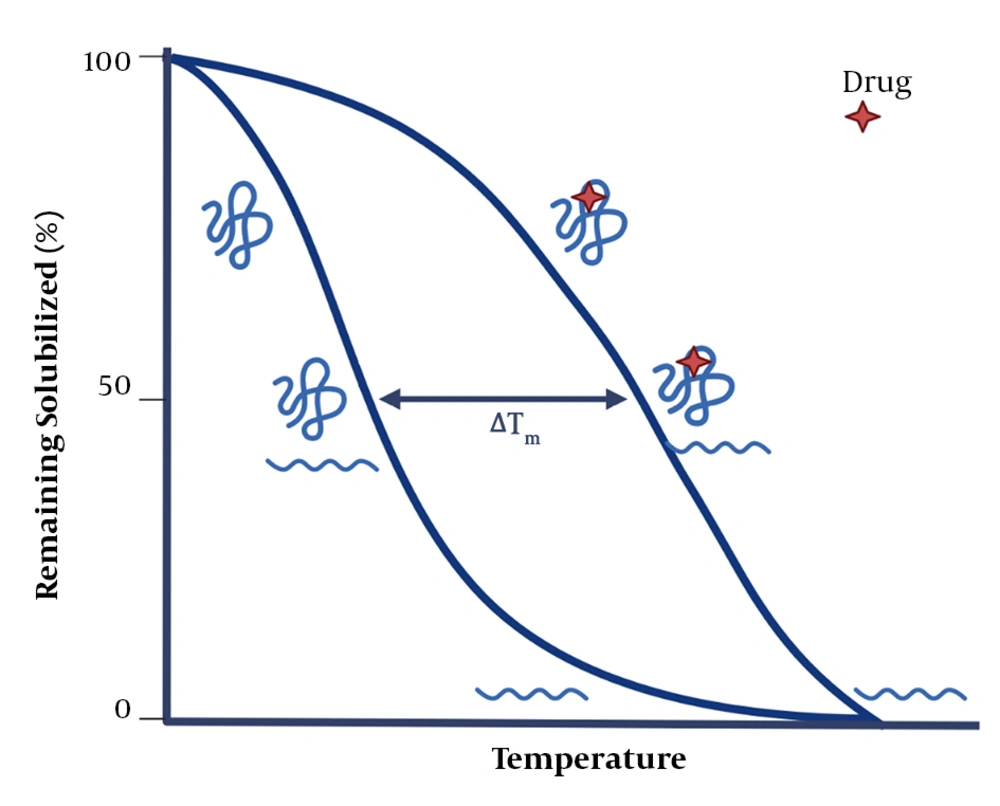
In the TPP technique, the CETSA technique is combined with mass spectrometry to more comprehensively study drug-protein interactions and to explore proteins’ intracellular thermal stability and proteolytic susceptibility. The TPP technique evaluates protein-ligand interactions directly in living tissues or cells and can detect post-translational changes. It should be noted that the ligand-protein complex becomes more stable against heat and chemical denaturation (Figure 4) (34-36, 50). Exposing proteins to thermal stress generally triggers the irreversible unfolding of proteins. As a result, proteins reveal their hydrophobic cores and subsequently aggregate (51-53). The temperature at which aggregations are formed (i.e., the melting temperature, Tm) increases in the presence of a ligand. So, a part of the energy supplied to the protein-ligand complex is used to separate the ligand from the protein (39, 54, 55). The evaluation of changes in the melting temperature (Tm) has been a valuable parameter in the pharmaceutical industry, allowing for creating compound libraries to find stabilizing ligands for proteins using fluorescence- or light-scattering–based techniques (56-59). In recent studies, the combination of Tm shift assays and the detection of stabilizing ligands for isolated targets via mass spectrometry have provided a broader detection power. Also, TPP can be used in living cells without a need for the labeling process, enabling researchers to conduct an unbiased investigation on drug targets. Only the TPP approach has combined all of these advantages (43).
The basics of the thermal shift assay (43). After drug-protein interaction, the stability of protein increases, requiring more energy for its denaturation.
For the first time, TPP was used to explore protein-protein interactions in 2014. This process is accomplished by providing ATP to living cells; the binding of ATP to proteins significantly stabilizes them against high temperatures. The stability of ATP-bound proteins can also be examined by the TPP method after adding ATP to cell extracts, confirming the accuracy of the findings (50).
In another study, several types of kinase inhibitors were used to investigate the efficacy of TPP. Kinases are enzymes that perform critical regulatory functions in cellular pathways. By TPP, it was possible to identify receptor-ligand interactions using a wide range of kinase inhibitors such as staurosporine and GSK3182571. These inhibitors changed kinases’ target proteins’ melting temperatures, as well as the thermal profiles of the regulatory subunits of the kinase complex (50).
Vemurafenib, which is used to treat Melanoma, causes severe photosensitivity and increases the levels of protoporphyrins (60). Based on the TPP method, it was revealed that after treating with vemurafenib, the main proteins affected were its cognate target, BRAF (61), and the off-target, ferrochelatase (FECH) (50). Ferrochelatase is dysregulated in protoporphyria, and its inhibition causes photosensitivity, a side effect of vemorafenib (62). Also, studies showed that alectinib, a second-generation anaplastic lymphoma kinase (ALK) inhibitor, was associated with photosensitivity (63), which also seems to be related to the effect of this drug on FECH. These studies proved that TPP could be used as a high-performance independent technique to study ligand-target interactions in cells.
Panobinostat, a histone deacetylase inhibitor, is an anticancer chemical substance used to treat diseases like myeloma and other malignancies (64). This drug has undesirable side effects such as hypothyroidism and seizures, indicating that it has off-target proteins (65). Recently, the TPP method was employed to explore these off-target proteins causing the drug’s side effects (41, 50). Since tyrosine is essential for the biosynthesis of thyroid hormones, suppressing phenylalanine hydroxylase reduces thyroid hormones’ production. On the other hand, it has been shown in various studies that tyrosine levels increase in tyrosinemia (66, 67). According to the results of a trial, panobinostat was suggested as an appropriate drug to treat tyrosinemia since it inhibits tyrosine synthesis by suppressing phenylalanine hydroxylase (36).
Drug repurposing is one of the essential applications of TPP (36). In addition to drug discovery, TPP can be used to analyze protein-protein interactions, as well as proteins’ functions and post-translational changes (68).
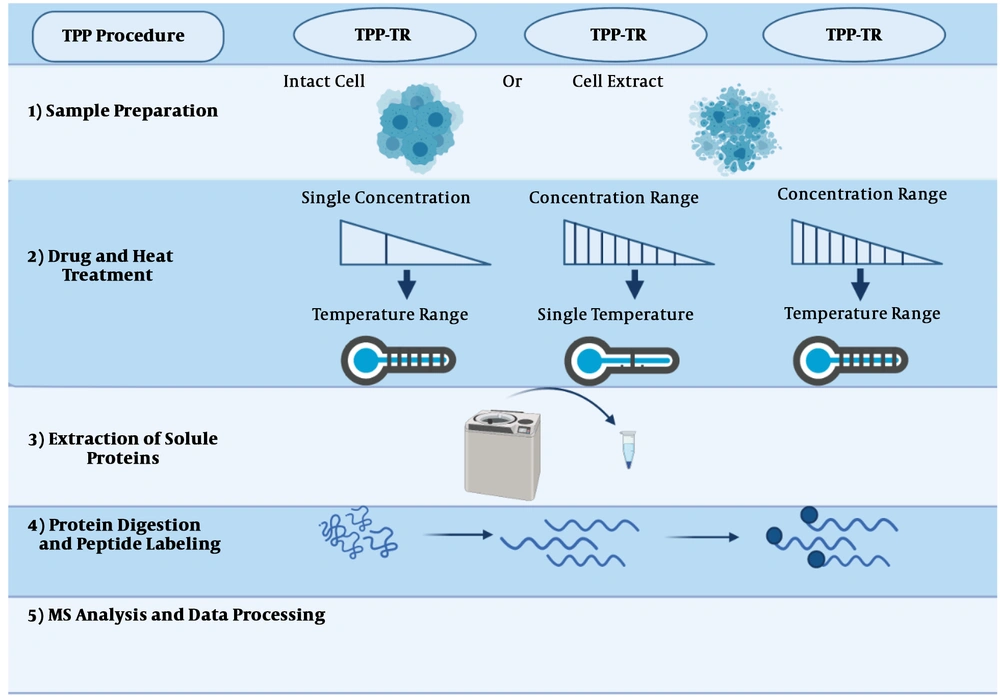
The protein thermal curve, which was generated and applied by the Savitski’s lab to purify proteins, was introduced as a strategy to detect drugs’ targets. The steps of this method have been illustrated in Figure 5 and will be described below.
The thermal proteome profiling (TPP) method can be conducted in each of the three modes: (1) compound concentration range (TPP-CCR); (2) temperature range (TPP-TR); and (3) two-dimensional TPP (2D-TPP). The general procedure has been described in (43).
2.5.1. Sample Preparation
Based on the purpose of the study, two distinct samples, intact cells, and cell extracts, are prepared for identifying indirect and direct targets, respectively. Using cell extracts, due to cell lysis and the dilution of cellular metabolites, cells’ normal metabolism is disrupted. As a result of cell explosion, the structure of cells falls apart, so only direct targets will be recognized. On the other hand, using intact cells, since cellular metabolism is preserved, upstream proteins can be recognized as indirect targets (Figure 5, step 1) (69).
2.5.2. Drug/Thermal Treatment
In this section, prepared samples are incubated with the desired drug at a temperature range (Figure 5, step 2). The selection of the drug concentrations is directly related to the next step, and just one dose or a concentration range can be used. When only one concentration is used, cells are exposed to a thermal range, called temperature range TPP (TPP-TR). In this approach, most of the drug’s targeted proteins can be identified. In the methods employing a concentration range (i.e., TPP-CCR), samples are treated with a concentration range of the combination at a unique temperature.
Recently, two-dimensional TPP has been developed, in which cells are examined at different drug concentrations and temperature ranges. This test provides the ability to designate the desired combination to effectively and immediately identify the target with high sensitivity. For instance, phenylalanine hydroxylase is an indirect target for panobinostat, which was not detectable by the TPP-TR method.
2.5.3. Extraction of Soluble Proteins
After treating cells with the drug and heat, they will be lysed, so denatured proteins can be removed using an ultracentrifuge (Figure 5, step 3) (34).
2.5.4. Protein Digestion and Peptide Labeling
At this stage, proteins are decomposed into peptides based on existing protocols (70). Then the isobaric tandem mass tags (TMT) method is applied for labeling. Before being subjected to mass spectrometry (MS), these labels have the same mass, but after MS, they are fragmented and yield reporter ions with different masses. These quantitative properties are compared between different experiments with similar MS conditions (Figure 5, step 4) (44).
2.5.5. MS Analysis and Data Processing
After MS analysis, the drug’s target proteins can be identified by comparing the control and treated samples. Most proteins are affected by ligand binding, but some target proteins do not show significant changes in their melting temperatures. By comparing the untreated and treated samples, the soluble proteins that have been affected by the drug can be easily recognized.
3. Conclusions
The drug discovery pipeline must respond quickly and effectively to the industry-wide challenges of enrollment rates and the pressures of strict regulatory requirements. Therefore, "omics" biology approaches are the most helpful tools in the target identification process. High-throughput proteomic technologies, which deliver a few thousand data points at just one run, have been expanded to drug discovery processes. Since the TPP method lies at the interface between proteomic and metabolic areas, it can help to answer several fundamental biological questions. The TPP method is used to solve many problems in drug discovery. Most notably, it facilitates the identification of the markers predicting drug efficacy and toxicity, provides an unbiased measurement of drug-target interactions and a rationale for understanding drugs’ adverse clinical outcomes, and suggests the repurposing of drugs for the treatment of other diseases (36).