1. Background
Rheumatoid arthritis (RA) is one of the most common autoimmune diseases. So far, no factor has been found to cause this disease. The prevalence of RA is about 1% in the human population worldwide; moreover, it is twice as prevalent in females than in males (1). A 10-year follow-up study from 2004 to 2014 in the United States showed that the prevalence of RA is increasing (2).
Rheumatoid arthritis is known as a complex disorder characterized by the inflammation of the synovium (the thin lining of a joint). Rheumatoid arthritis is a chronic disease in which genetic and environmental factors contribute to decreasing the tolerance to self-antigens (3). TNF-α is an inflammatory cytokine that plays an important role in the development of RA (4, 5). TNF-α initiates and regulates the cytokine cascade during the pathogenesis of RA. TNF-α stimulates monocytes/macrophages, fibroblasts, and endothelial cells to produce IL-l and IL-6 or chemokines (CXCL8, CCL2), which are heavily involved in the pathogenesis of RA including leukocytes infiltration and tissue destruction. Inflammation induced by IL-1 and TNF-α can lead to the increased expression of the matrix metalloproteinase, collagenase, and elastase during the pathogenesis of RA, which is responsible for cartilage degradation and bone resorption (6, 7).
TNF-α affects the target cell via two receptors, TNF-RI (TNFRSF1A) and TNF-RII (TNFRSF1B). Although the homology of the extracellular domains of the TNF-α receptor is very close, their intracellular domains are different (8). TNF binding to TNF-RI triggers apoptosis while its binding to TNF-RII triggers cell survival. Both TNF-RI and TNF-RII are expressed in the synovial tissue in patients with RA (7, 9). The TNF-RI and TNF-RII genes are located on chromosomes 12p13 and 1p36, respectively (10).
Genome-wide association studies have identified that 1p36 locus is associated with RA. Interestingly, TNF-RII is located within this gene area with 10 exons (11). Reports indicate that one of the most important single nucleotide polymorphisms is located in exon 6. This missense mutation results in methionine to arginine substitution (ATG → AGG) at position 196 (196R), which is within the fourth extracellular domain of TNF-RII (12).
Based on the biological and molecular analysis, the TNF-RII gene is considered as one of the best candidates among RA-susceptibility genes. SNP analyses at 196 (rs1061622) indicate that the 196R mutant allele has a different performance than the 196M wild-type allele (13). As reported, there appears to be an association between the exon 6 polymorphism (T676G) and susceptibility to RA (14).
Genetic markers may differ among different populations. These differences may affect the TNF-RII association with RA disease in different populations, leading to contradictory results. Previous studies have reported an association between TNF-RII 196R allele polymorphisms and RA in other populations (15, 16). Therefore, we need to evaluate the genotype and allele frequency in 196 (rs1061622) the TNF-RII gene and investigate the genetic risk of this polymorphism in our population.
2. Objectives
In this study, we tried to evaluate the association between single nucleotide polymorphisms (SNPs) of the TNF-RII gene (rs 106 16 22) and the risk of susceptibility to RA in a sample of the Iranian population.
3. Methods
3.1. Participants
After confirming the protocol of the study by the Ethics Committee of the Semnan University of Medical Sciences (ethical code; 92/372650), the study was conducted in the Department of Immunology at the University. All patients entering the study signed informed consent forms. Blood samples were taken from RA patients and healthy controls and the genomic DNA of all individuals was extracted (Qiagen DNA mini kit, Basel) from peripheral blood cells.
Our case-control study included 100 RA patients. All patients were screened as RA patients according to the American College of Rheumatology criteria. Patients were selected from those who referred to Tehran Baqiyatallah Hospital for RA treatment. The control group consisted of 100 healthy individuals.
3.2. DNA Extraction
Using QIAamp® DNA Mini extracting kit, genomic DNA was extracted from 500 µL of blood lymphocytes; the extracted DNA was stored at -20ºC until use for genotyping.
3.3. SNP Genotyping
The real-time PCR method was used for the determination of M196R allele polymorphism in the TNF-RII gen. All PCR tests were performed in a volume of 50 µL, containing TaqMan Universal PCR Master Mix, specific TaqMan SNP genotyping assays, life technologies, assay ID (C 8861232 20, (part number) 4351379, USA) and genomic DNA.
Thermal cycling conditions were 5 min at 95ºC, followed by 40 cycles of 95ºC for 15 s, and annealing and extension were performed at 60ºC for one minute. After PCR, to measure the allele-specific fluorescence, the genotyping of rs061622 SNPs was performed by TaqMan allelic discrimination with a thermocycler 7900 ABI system (17).
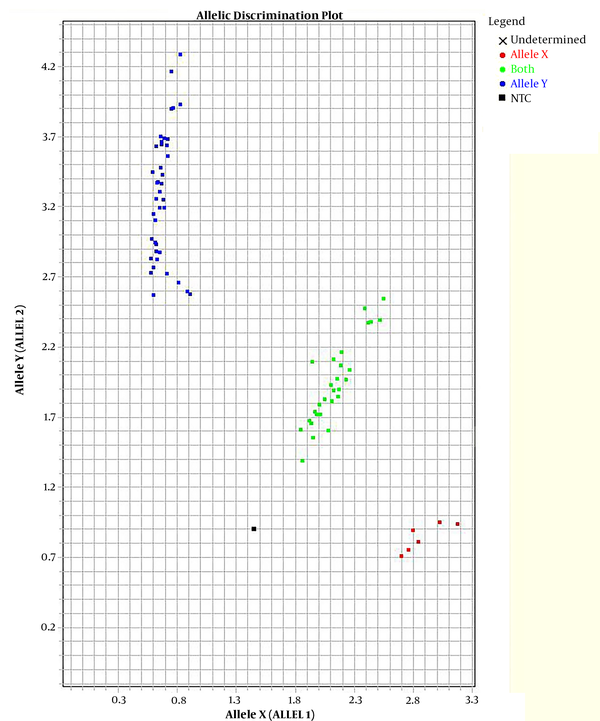
In this study, two TaqMan probes labeled with different fluorescent dyes were used for each sample in an allelic discrimination assay. The probes labeled with fluorescent dye FAM had a perfect match with the wild-type allele. The probes were labeled with fluorescent dye VIC because it was perfectly matched with the mutated allele (Figure 1).
TaqMan™ genotyping plot; each sample was analyzed with two probes, one specific for the wild-type and one for the mutation. The strength of fluorescence from each probe was plotted on a graph (wild-type on X-axis, mutant on Y-axis). Each sample is represented by a single point. The samples fall into three clusters representing the possible genotypes: homozygous wild-type (blue), homozygous mutant (red), heterozygous (green).
3.4. Statistical Analysis
We used SPSS version 22 software for statistical analyses. The t-test was used to compare the mean age of cases and controls. In addition, Hardy-Weinberg equilibrium (HWE) was investigated in case and control groups of RA. The chi-square test was used to compare genotype frequencies and allele and genotype distributions in patients and controls. The associations between genotypes of the TNF-RII gene and RA were assessed by computing odds ratios (ORs) and 95% confidence intervals (95% CIs) using logistic regression analysis. The significance level was set at P < 0.05.
4. Results
Our study aimed to investigate the association between the functional 196R polymorphism of TNF-RII and RA. The mean age (± SD) of the participants was 48.3 ± 13.1 years, ranging from 23 to 77 years. The mean age (± SD) of patients was 48.1 ± 12.6 years. Table 1 presents the characteristics of RA patients. The differences between normal and patient groups were not significant in terms of age and sex (P > 0.05, Table 1).
Table 2 presents the genotype and allele frequencies of non-synonymous polymorphism rs1061622 of the TNF-RII gene in RA patients and controls. Significant differences were observed between genotype frequencies of the two groups in terms of 196R polymorphism. Moreover, the rs1061622 variant increased the risk of RA. The GG genotype was more common in RA patients than in normal subjects (OR for GG genotype: 2.6, 95% CI: 1.135 - 5.6, P = 0.035) (Table 2). Moreover, the G allele frequency in RA patients the rs1061622 was significantly higher than in the control group (OR for G phenotype: 2.30, 95% CI: 1.1 - 5.1, P = 0.04) (Table 2).
| Genotype | RA Patients | Control | OR | Confidence Interval | P |
|---|---|---|---|---|---|
| 196 MM | 47 (47) | 53 (53) | 0.9 | (0.5 - 1.8) | 0.74 |
| 196 MR | 41 (41) | 42 (42) | 2.6 | (1.135 - 5.6) | 0.035 |
| 196RR | 12 (12) | 5 (5) | |||
| Allele196M | 135 (67.50) | 148 (74) | 2.3 | (1.1 - 5.1) | 0.04 |
| 196R | 65 | 52 |
Genotype and Allele Frequency Distribution of TNF-RII rs1061622 Polymorphism in Rheumatoid Arthritis (RA) Patients and Healthy Subjectsa
Of a total of 135 (32.5%) alleles in patients with RA, 65 cases had 196R alleles, whereas of a total of 200 (26%) alleles in healthy individuals, 52 cases had 196R alleles. In addition, the G allele increased the risk of RA.
5. Discussion
In this case-control study, we investigated the relationship between TNF-RII rs1061622 polymorphism and RA in the Iranian population. The results of the present study indicated a significant relationship between TNF-RII rs1061622 polymorphism and RA disease. There seems to be a positive correlation between TNF-RII rs1061622 polymorphism and RA.
The results showed that the TNF-RII R allele was present in 32.5% and 26% of RA patients and controls, respectively. The association between the polymorphism and the disease was confirmed by the odd ratio (CI = 95%, OR = 2.6, P = 0.035). Several studies have been conducted in different populations to investigate the relationship between TNF-RII polymorphism and RA disease. The findings of this study are in line with the results of Barton et al. studies, which demonstrated an association between the TNF-RII 196R/R genotype and familial RA in the UK and among French Caucasian populations (14, 15), and Dieude et al. study in the Japanese population (16). However, studies conducted on the Swedish population did not show any significant association between RA and the TNF-RII 196M/R polymorphism (18). It seems that heterozygous patients carrying the TNF-RII 196R allele show RA at an earlier age than earlier age than those carrying homozygous TNF-RII 196M allele. The age of the disease onset is an important factor in the study of RA. The study by Ghelani et al. on RA patients in Southeast Asia did not show any association between TNF-RII 196R/R genotype and RA disease (19).
Several studies investigating the relationship between the TNF-RII 196M/R polymorphism and RA severity have reported controversial results (19-21). van der Helm-van Mil et al. (22) and Glossop et al. (20) studies did not show any significant association between the severity of RA disease and 196R allele (19, 20). However, Goeb et al. study revealed a significant correlation between TNF-RII 196R allele and radiographic severity and diagnosis of RA (21, 23).
Previous studies have shown that the TNF-RII 196 M/R gene tends to characterize increased cytokine production and apoptosis after TNF-α stimulation (13, 24). Studies by Horiuchi et al. showed that a polymorphism in codon 196 exon 6 TNF-RII changes methionine to arginine which increases IL-6 production.in the cells carrying the 196R allele (13). Till et al. showed that the change of methionine to arginine changes the pathway for TNF-RII apoptosis that is performed by NF-kb signaling (24).
A recent meta-analysis study performed by Song et al. (25) showed an association between the functional TNF-RII 196M/R polymorphism and RA in the European population and East Asian population. In addition, in previous studies, a significant association was found between TNF-RII 196R polymorphism and lupus disease in the Japanese population (26). As noted, this association between RA and TNF-RII polymorphism was studied in the Japanese population. A similar study in France showed the same results (15).
In conclusion, we investigated the association between TNF-RII gene (rs1061622) polymorphisms and RA in a sample of the Iranian population for the first time. Our results supported a significant association between the missense mutation, which involves a single base substitution at codon 196 (ATG → AGG) in exon 6 of the TNF-RII gene, and susceptibility to RA.
Association studies may be limited by the heterogeneity of the population, small sample sizes, and the statistically significant differences between experimental and control groups. The difference in results could be due to different patient population genetic background in different studies. To confirm our findings, it is necessary to conduct further association studies in different ethnicities.
.jpg)