1. Background
Focal segmental glomerulosclerosis (FSGS) is a type of nephrotic syndrome which is diagnosed using renal biopsy and may be found as either primary or secondary conditions. Primary FSGS occurs without an identifiable cause and the secondary type occurs in response to previous glomerular injury, glomerular hypertension, or hypertrophy (1). While the clinical presentation of FSGS is often heterogeneous, a characteristic feature of the disease is proteinuria due to loss of filtration barrier of glomeruli (2). Histological characteristics of FSGS also include scattered sclerosis of glomeruli in which only a segment of the capillary is affected (3). It is the most common cause of acquired chronic renal insufficiency in children and frequently leads to progression to the end-stage kidney disease (ESKD) (4). Factors that appear to mainly affect the prognosis include the degree of proteinuria and renal dysfunction, histologic findings, and the response to therapy. In the present study we focused on the discovery of urinary proteins responsible for developing more severe renal dysfunction as a prognostic factor using proteomics tools.
2. Objectives
In the present study, we attempted to discover urinary excreted proteins which can be used for the differentiation of patients with good and bad prognosis. These non-invasive biomarker candidates would be useful in the follow-up and detection of disease progression without using biopsy.
3. Patients and Methods
Second morning urine samples were collected from 11 patients with biopsy proven FSGS (male = 7, female = 4, mean age = 36.36), at Labbafinejad Hospital during 2011. For each patient eGFR was calculated by CKD-EPI equation at presentation. In order to study differential proteins among good and bad prognosis patients, we categorized all patients based on eGFR. Since more severe renal dysfunction at presentation is generally associated with poor renal survival (5), five patients considered as mild disease state (eGFR > 60 cc/min/1.73 m2) and five patients with advanced disease state and worse prognosis (eGFR < 60 cc/min/1.73 m2) were enrolled. Urine samples were concentrated and desalted with ultrafiltration (Millipore, Billerica, MA, USA with a 3 kDa cut off) and then were treated with acetone (up to 80% v/v), dried and re-suspended in 0.1 M ammonium acetate (pH 5). Protein concentration was then determined using the BCA (bicinchoninicacid) protein assay (Pierce, Thermo Scientific, USA) and proteins were further digested by trypsin. Digested peptides were suspended in an appropriate buffer and injected to a liquid chromatography tandem mass spectrometry (nLC-MS/MS) coupled online to a Q Exactive mass spectrometer (both-Thermo Scientific, Bremen, Germany). For details of the sample preparation protocol, MS analysis and label-free quantification procedure see the article of Kalantari et al. (6). Protein profiles then were analyzed using supervised multivariate statistical analysis. Patients were categorized based on GFR (renal dysfunction). A predictive model was then constructed and validated by 7 fold cross-validation and significant proteins were determined. Gene-set enrichment and pathway analysis on significant proteins were performed using “DAVID” software.
4. Results
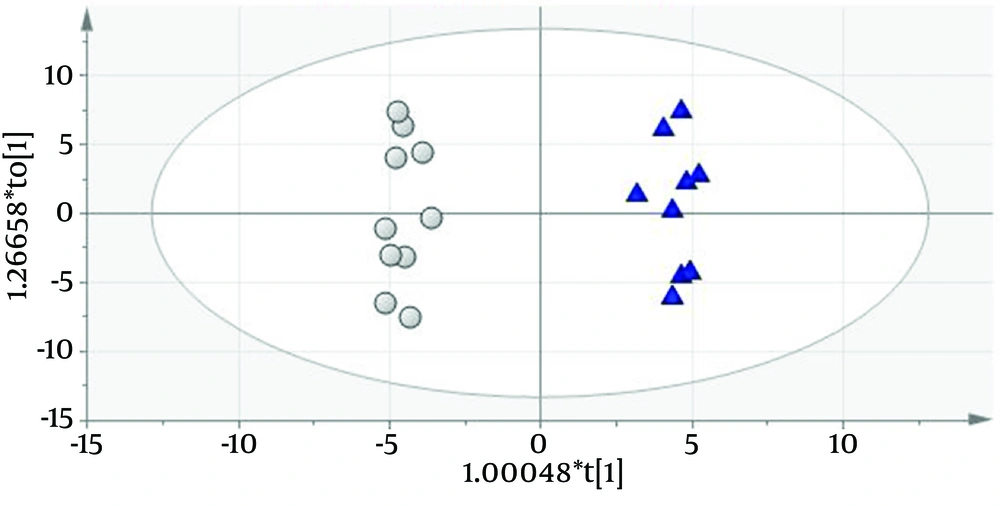
Urinary protein profiles of FSGS patients using 110 protein entries were quantified as described by Kalantari et al. (7). Differential proteins between the two groups with different prognosis features (eGFR < 60 and > 60 cc/min/1.73 m2) were obtained using orthogonal projection to latent structures discriminant analysis (OPLS-DA) (8). A predictive model was constructed by this method (Q2 = 0.861 and R2 = 0.619) which had 100% accuracy (Figure 1). Fifty four significant biomarkers were obtained from the predictive model of which top twelve (six most positively and six most negatively correlating with GFR) are described in Table 1 as putative FSGS progression biomarkers.
| Protein ID | Protein Name | Biological Process | Cellular Component | Molecular Function | Fold Change (Low GFR/high GFR) | Up/Down Regulation |
|---|---|---|---|---|---|---|
| RNAS2 | Ribonuclease 2 | RNA catabolic process | Extracellular region/lysosome | Ribonuclease activity | 7.32 | ↑ |
| CD59 | CD59 glycoprotein | Negative regulation of activation of membrane attack complex | Anchored to external side of plasma membrane/extra cellular space | Potent inhibitor of the complement membrane attack complex (MAC) action | 7.21 | ↑ |
| PTGDS | Prostaglandin-H2 D-isomerase | Prostaglandin biosynthesis/Lipid biosynthesis | Extracellular space/Golgi apparatus/rough endoplasmic reticulum | Fatty acid binding/prostaglandin-D synthase activity | 5.81 | ↑ |
| B2MG | Beta-2-microglobulin | Regulation of immune response | Extracellular space/MHC class I protein complex | Involved in the presentation of peptide antigens to the immune system | 4.98 | ↑ |
| AMBP | Alpha-1-microgolbulin | Negative regulation of immune response | Extracellular space/ plasma membrane | Serine-type endopeptidase inhibitor activity | 4.39 | ↑ |
| SULF2 | Extracellular sulfatase Sulf-2 | Glomerular basement membrane development | Extracellular space/plasma membrane | N-acetylglucosamine-6-sulfatase activity/calcium ion binding | 4.33 | ↑ |
| CBG | Corticosteroid-binding globulin | Glucocorticoid metabolic process/regulation of proteolysis | Extracellular space | Serine-type endopeptidase inhibitor activity/steroid binding | 1.61 | ↓ |
| AFAM | Afamin | Vitamin transport | Extracellular space | Vitamin E binding | 1.64 | ↓ |
| MXRA8 | Matrix-remodeling-associated protein 8 | Fibrosis process | Membrane | May play a role in the maturation and maintenance of blood-brain barrier | 1.71 | ↓ |
| CO6A1 | Collagen alpha-1(VI) chain | Cell adhesion/extracellular matrix disassembly | Endoplasmic reticulum lumen/sarcolemma | Platelet-derived growth factor binding | 1.74 | ↓ |
| ACTG | Actin, cytoplasmic 2 | Innate immune response/adherens junction organization | Cytoskeleton/extracellular vesicular exosome | ATP binding/structural constituent of cytoskeleton | 1.76 | ↓ |
| HPT | Haptoglobin | acute-phase response/positive regulation of cell death/response to hydrogen peroxide | extracellular space | antioxidant activity/catalytic activity | 2.26 | ↓ |
5. Discussion
Prognosis is important to patients, clinicians, public health, and health policy makers and glomerular filtration rate (GFR) is one of the effective prognostic factors for patients with declining renal function (9, 10) and glomerular diseases. Urinary biomarkers derived from a predictive model which reflect the prognosis of glomerular diseases (based on GFR in the current study) could be considered as useful noninvasive markers for rapid, reliable and accurate diagnosis and monitoring the progression of the disease in comparison with current traditional invasive approaches, however the causes of up/down regulation of these candidates is not clear and need further experiments. Some of the most significant biomarker candidates are presented here:
RNAS2 had the greatest-fold change (7.32) as the overrepresented biomarker in patients with worse prognosis (eGFR < 60 cc/min/1.73 m2). A strong correlation between serum RNase levels and renal insufficiency was previously reported by Humphrey et al. (11). RNAS2 is a 3kDa protein which is found in body fluids (including urine) (12) and some tissues. The pathologic reason of its urinary elevation in glomerulosclerosis is not clear; however, to the best of our knowledge, it is reported here for the first time as a prognostic candidate marker for FSGS.
HPT (haptoglobin) is reported here as the underrepresented biomarker for FSGS progression with the greatest fold change (2.26). An association between the haptoglobin genotypes and renal function decline in individuals with long-standing type 1 diabetes was previously reported by Costacou et al. (13). The exact role of haptoglobin in progression of FSGS is not well defined and a wide targeted genomic and proteomic experimental design is required for this purpose. In the current study, some of the presented biomarkers have reported before as already known proteins implicated in glomerular disease including: B2MG (14) and AMBP (15), AFAM (6), but most of the other candidates identified in this study are novel. Gene-set enrichment analysis by “DAVID” software (16) resulted in identification of eight significant biological processes of which “acute inflammatory response” (P = 5.3 × 10-7), “blood coagulation” (P = 2.7 × 10-4) and “regulation of homeostatic process” (P = 5.3 × 10-3) are the more specific processes relevant to disease progression. Two of the proteins which were in the panel list (composed of 50 proteins) and were enriched in all eight significant processes were A1AG1 (orosomucoid 1) and THRB (prothrombin). We suggest these two proteins are s important proteins implicated in the pathogenesis of FSGS. Pathway analysis against KEGG database using “DAVID” software resulted in two single significant pathways which were also shown to be enriched in the previous study from our group (7) which are the complement and coagulation cascades (P = 8.2 × 10-5). It may suggest that impairment of complement pathway regardless of the criterion for judgment prognosis determination (either GFR or responsiveness to steroids) plays an important role in the progression and pathogenesis of FSGS. Impairment of this pathway in FSGS progression was consistent with our previous findings in which prognosis was determined based on the response to therapy.
In conclusion, a panel of urinary prognostic biomarkers was reported for FSGS. The most significant over- and underrepresented proteins in patients with worse prognosis in comparison with patients with a better prognosis were RNAS2 and HPT, respectively. These candidates were obtained from a predictive model which clustered patients based on GFR. In conclusion, involvement of proteins responsible for acute inflammatory response and also involvement of complement and coagulation pathways in disease progression were confirmed in our study using bioinformatics methods.