1. Context
Contact tracing is a principal public health practice for containing further propagation of the virus through limiting contacts between infected cases and persons adjacent to them (eg, family members, health care providers, healthcare personnel, etc.) (1-3). Contact tracing is principally significant for the COVID-19 outbreak, where a large number of carriers are silent, pre-symptomatic, or may present only mild symptoms and are thus usually not tested, despite having the potential to promulgate the disease (4). In the context of COVID-19, contact tracing is a public health response to detect and inform those individuals who may have been in close contact with an infected person every day for two weeks (5, 6). Accordingly, if an individual is confirmed positive for COVID-19, every other individual who had possibly been in close contact is tracked and recommended to go into protective self-quarantine for cutting off the transmission chain of the disease in the community (7).
To overcome the limitations of traditional contact tracing, digital-based contact tracing has been adopted (8). One promising type of digital contact tracing is the implementation of mobile-based contact tracing applications (apps). Such apps use mobile devices to promptly detect and alert users who may be in close contact with a confirmed-COVID-19 case (9). Due to the wide accessibility and affordability of mobile devices, employing mobile-based contact tracing apps can lead to making the public health process of contact tracing more efficient on a massive scale (10).
Mobile-based contact tracing systems offer a practical solution to controlling the spread of COVID-19; however, standardized data collection as one of the designing specification criteria to achieve a uniform and mass tracing app acceptance is a great challenge (11, 12). Moreover, from a data management perspective, the novelty of COVID-19 has created major gaps in data harmonization, integration, and unified reporting of disease as a basis for investigating many unfamiliar clinical aspects and outcomes of the disease, characterizing the public health threat, and supporting health authorities’ decisions (13).
The human-to-human spread of COVID-19 requires active case identification, that is, early confinement, timely testing, and treatment, besides detection and future tracking of persons who may be in close contact with infected cases (14). Meanwhile, a large number of reports inflowing the health care systems from varied networks and formats need to be validated. Current surveillance systems are generally not constructed to meet such data requirements. Moreover, vagueness and postponement of surveillance data due to isolated and heterogeneous health information systems are a barrier to data exchange among these systems, which have led to limited consistency of epidemiologic studies (15).
To our knowledge, no comprehensive data collection template currently exists that has been designed to capture high-quality, consistent, and standardized data regarding COVID-19 contact tracing.
2. Objectives
To address this priority, the current study aims to determine a minimum dataset (MDS) as an essential measure before the design and implementation of a digital contact tracing system. Accordingly, we sought to develop a COVID-19 Contact Tracing Minimal Dataset (COV-CT-MDS) based on mobile devices due to their ability to appropriately document contact tracing data during COVID-19.
3. Methods
This was a cross-sectional study conducted in 2020 following a combination of literature review and a broad discussion with a multidisciplinary team of involved healthcare experts, as follows.
3.1. Literature Review
3.1.1. Search Strategy
A systematic review was undertaken to extract the primary data elements to include in COV-CT-MDS. This systematic review was reported according to the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (16). PubMed, Scopus, Web of Science (WOS), Science Direct, and ProQuest databases were reviewed between 1 January 2020 and 20 December 2020 to determine the required data elements, features, and attributes for designing a mobile-based COV-CT-MDS. The following search terms were used (designed using English MeSH keywords) to maximize the output from literature findings: [COVID-19 OR Novel coronavirus OR SARS-CoV-2 OR n-CoV2] AND [Mobile phone OR Smartphone OR Cell phone OR Mobile Apps OR Mobile health] AND [Contact tracing OR Contact tracking].
3.1.2. Study Selection
Two independent researchers (M: SH and H: K-A) reviewed the titles and abstracts of the articles extracted from the initial search, and then full-text articles were obtained for detailed evaluation. Finally, we read the full text of articles and recognized potentially eligible studies to be included in the systematic review.
The following criteria were considered as the inclusion criteria:
(1) Type of a study: Original or review research papers were selected, and newspapers, reports, editorial, letters, posters, and conference papers were not examined.
(2) Date of publication: Papers published between 1 January 2020 to 20 December 2020
(3) Language: English language
(4) Text availability: Full-text papers with the keywords in the title or abstracts
(5) Content analysis: At least two of the following reporting parameters: (1) basic/general, (2) clinical, (3) para-clinical, (4) geo-locational, and (5) contact/exposure data classes.
Finally, the probable data elements to be included in the COV-CT-MDS were recorded in a checklist with two administrative and clinical sections.
3.1.3. Data Extraction
For each eligible research, the following information was extracted based on a designed data extraction form, which included the first author, country, year of publication, study design, and reporting data classes in the two non-clinical and clinical data categories. The results were organized under the following categories: (1) data categories, (2) data classes, (3) data fields, and (4) data features and attributes.
3.2. Delphi Technique
3.2.1. Questionnaire Design
After conducting the necessary literature review and receiving expert advice, we developed a questionnaire. We invited 20 experts, including five infectious diseases specialists, five virologists, five health information management (HIM), and five clinical epidemiologists, in a two-round Delphi survey. The questionnaire included the following parts: (1) demographical data, (2) clinical finding, (3) geolocation location, (4) relocation data, and (5) contact/ exposure data.
3.2.2. Data Analysis
The experts participating in the study were asked to score the tabulated data elements in terms of their importance using a five-point Likert scale (ranging from 1: “very slightly important” to 5: “highly important”). Data fields with less than 50% agreement were excluded in the first round, while those with greater than 75% agreement were included in the primary round. Those with 50% to 75% agreement were surveyed in the second round, and if there was 75% consensus over a subject, it was regarded as a final data field.
4. Results
4.1. Characteristics of Included Studies
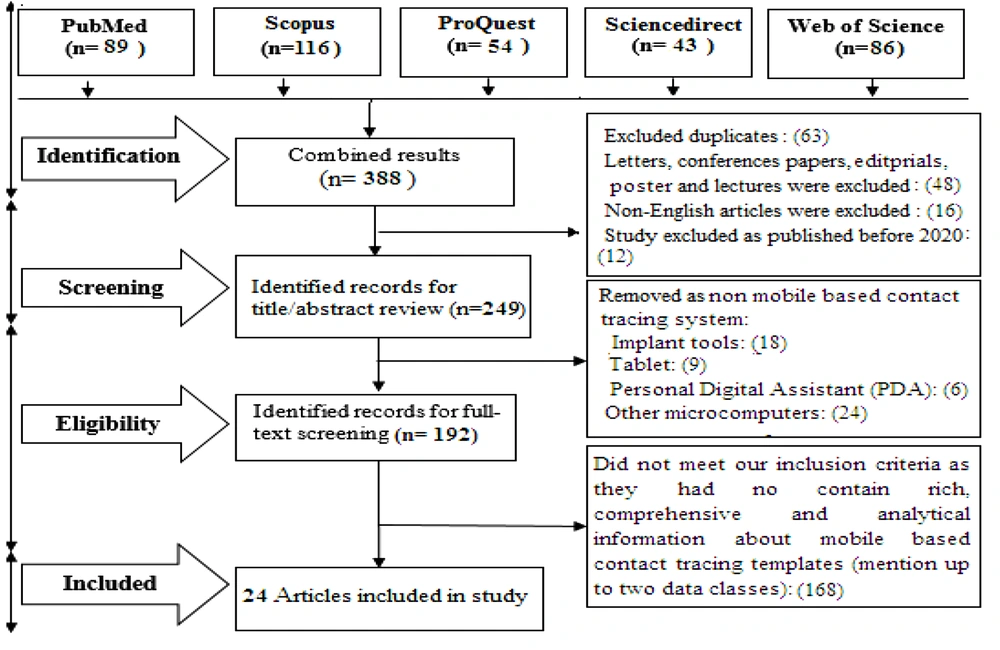
A total of 388 articles were retrieved from the literature search. After the removal of duplicate articles and those not meeting the inclusion criteria, 24 articles that satisfied all the inclusion criteria were included in the analysis. Figure 1 summarizes the selection process (PRISMA chart).
4.2. Identifying the Proposed Data Field
The proposed data fields after the literature review were divided into administrative and clinical data sections, nine data classes, and 198 data fields (Table 1).
| First Author (2020) | Method | Data Classes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Administrative | Clinical | ||||||||||
| Basic | Geolocation | Occupational | Relocation | Contact | Exposure | Clinical | Manifestations | Vital Signs | Referral | ||
| Bassi et al. (17) | Descriptive | * | * | * | |||||||
| Basu (18) | Case study | * | * | * | |||||||
| Davalbhakta et al. (19) | Review | * | * | * | * | ||||||
| Ekong et al. (20) | Exploratory review | * | * | * | * | ||||||
| Hassandoust et al. (21) | Developmental | * | * | * | * | * | |||||
| Martin et al. (5) | Review | * | * | ||||||||
| Parker et al. (22) | Descriptive | * | * | * | |||||||
| Rahman et al. (23) | Case study | * | * | ||||||||
| Shubina et al. (24) | Retrospective | * | * | ||||||||
| Vuokko et al. (25) | Descriptive | * | * | * | |||||||
| Prabu et al. (26) | Exploratory review | * | * | * | |||||||
| Teixeira and Doetsch(27) | Descriptive | * | * | * | |||||||
| Kondylakis et al. (28) | Review | * | * | * | * | ||||||
| Nakamoto et al. (29) | Developmental | * | * | * | * | ||||||
| Altmann et al. (30) | Retrospective | * | * | ||||||||
| Dar et al. (31) | Developmental | * | * | * | |||||||
| Singh et al. (32) | Review | * | * | ||||||||
| Urbaczewski and Lee(33) | Retrospective | * | * | * | * | ||||||
| Whaiduzzaman et al. (34) | Developmental | * | * | * | * | * | |||||
| Bianconi et al. (35) | Descriptive | * | * | * | * | ||||||
| Grantz et al. (36) | Prospective | * | |||||||||
| Ming et al. (9) | Retrospective | * | * | * | * | * | |||||
| Wirth et al. (37) | Scoping review | * | * | * | * | ||||||
| Nijsingh et al. (10) | Descriptive | * | * | * | |||||||
Several data fields were excluded after the second round of Delphi. Thus, of the 198 proposed data fields, 117 fields were excluded from the study, and 81 data fields were finalized (Table 2).
| Decision | Agreement Rate (%) | Frequency |
|---|---|---|
| First Round | ||
| Inclusion | < 75 | 58 |
| Exclusion | > 50 | 92 |
| Entering in second round | 50 - 75 | 48 |
| Second Round | ||
| Inclusion | < 75 | 25 |
| Exclusion | > 75 | 23 |
The final reporting template is composed of two data sections, nine data classes, and 81 data fields. Table 3 lists the data sections, classes, fields, their formats and values, and corresponding reference SNOMED-CT codes.
| Data Element | Feature Content | Feature Format | SNOMED-CT Category | SNOMED-CT Codes |
|---|---|---|---|---|
| General Characteristics | ||||
| Full name (11-16, 34, 36, 38, 39) | String | Observable entity | 371484003 | |
| Age (5-9, 12, 14, 18, 34, 38, 40, 41) | Forced choice | Qualifier value | 764868004 | |
| Gender (2, 4, 9, 10, 12, 15, 21, 36, 40, 41) | M: 1 F: 0 | Binary | Clinical finding | 703118005 |
| National ID number (4, 6, 9, 13, 18, 34, 40) | xxx- xxxxxx-x | Numerical | Observable entity | 422549004 |
| Citizenship (2, 15, 18, 21, 34, 36, 39, 41) | Iranian; Non-Iranian | Binary | Social concept | 275595001 |
| Medical record number (4, 5, 7, 12, 13, 16, 18, 21, 31, 40) | xx-xx-xx | Numerical | Observable entity | 398225001 |
| Level of education (5, 10, 12, 13, 15, 18, 36, 39, 41) | Primary; Secondary; Tertiary | Forced choice | Observable entity | 224300008 |
| Marital status (5, 7, 9, 12, 13, 16, 18, 34, 36, 39, 40) | Single; Married; Widow; Other | Forced choice | Clinical finding | 87915002 |
| Monthly income (3, 5, 6, 9, 13, 16, 21, 34, 36, 39, 41) | Low: < 120$; Medium: 120$ - 250$; High: > 250$ | Forced choice | Clinical finding | 424860001 |
| Family relationship to index cases (5, 6, 9, 15, 34) | Nuclear family; Extended family | Binary | Social concept | 394568007 |
| Phone number (4-6, 9, 13, 15, 16, 40) | +98 xxx xxx xxxx | Numerical | Observable entity | 398198004 |
| Healthcare facility unique ID (5, 6, 15, 18, 21, 40) | xxxxx | Numerical | Observable entity | 713578002 |
| Frontline health worker ID (4-6, 9, 10, 13, 15, 16, 36) | xxxxx | Numerical | Observable entity | 713578002 |
| Relationship with the source case (5, 6, 12, 14, 15, 18, 21, 34, 40, 41) | Partner / spouse; Family member; Other | Forced choice | Clinical finding | 852071000000103 |
| Geolocation Data | ||||
| Place of birth (6, 14, 15, 18, 21, 40, 41) | Geographical location: Province, city, village | String | Environment/ location | 315446000 |
| Resident situation (5, 6, 8, 15, 40) | Tenant; Owner; Other | Forced choice | Environment/ location | 184097001 |
| Residential address (3, 4, 6, 8, 9, 14, 36, 40) | String | Observable entity | 433178008 | |
| Postal code / zip code (3, 6, 8-10, 15, 36, 41) | xxxxx-xxxxx | Numerical | observable entity | 184097001 |
| Place of contact (3-6, 10, 11, 14-16, 36, 41) | Workplace; Home; Public place; Other; Unknown | Forced choice | Environment/ location | 257710009 |
| Location case identified (4, 6, 11, 13, 15, 21, 40) | Geographical location | String | Environment/ location | 706956001 |
| Origin of travel (5, 6, 15, 16, 34, 41) | Geographical location | String | Environment/ location | 224803003 |
| Travel destination (6, 10, 13, 16, 36, 39) | Geographical location | String | Environment/ location | 224807002 |
| Address of healthcare organization (3, 6, 13, 21, 39-41) | String | Observable entity | 184097001 | |
| Isolation/quarantine location (6, 13, 16, 40) | Self-isolation at home; Hospital; Long term care facilities; Other | Forced choice | Procedure | 1321131000000109 |
| Clinical Characteristic | ||||
| Symptom incidence (3, 4, 6, 7, 10, 12, 16, 31, 39, 40) | Asymptomatic; Pre symptomatic | Forced choice | Qualifier value | 264931009 |
| Date of symptom onset (6, 9, 12, 13, 15, 31, 36, 39, 40) | yyyy /mm/ dd | Integer | Observable entity | 520191000000103 |
| Days from exposure to symptom onset (8-10, 12, 13, 15, 36, 38, 39) | xx | Numerical | Qualifier value | 307474000 |
| Days from illness onset to first admission (5, 6, 10, 12, 15, 18, 38) | xx | Numerical | Qualifier value | 307474000 |
| Days from diagnosis to treatment (8, 12, 14, 40) | xx | Numerical | Qualifier value | 432213005 |
| Date of diagnosis (10, 12, 14, 18, 40, 41) | yyyy /mm/ dd | Integer | Observable entity | 432213005 |
| Covid-19 classification (10, 12, 18) | Confirmed; Probable; Unknown | Forced choice | Situation | 395098000 |
| Covid-19 status (9, 14, 18) | Active; Inactive; Recovered | Forced choice | Clinical finding | 110278006 |
| Case finding approaches | Random screening; Symptomatic case referral; Contact tracing; Other | Forced choice | Clinical finding | Country, Province/ State, City, |
| Prior hospitalization (3, 5, 9, 10, 12, 14, 34, 38) | Yes; No | Numerical | Clinical finding | 314503007 |
| Self Reported Clinical Manifestation | ||||
| Fever/chill (4-7, 10, 13, 18, 21, 31, 40) | Yes; No | Binary | Qualifier value | 14732006 |
| Cough (4, 6, 9, 10, 13, 18, 41) | Yes; No | Binary | Clinical finding | 314503007 |
| Dyspnea (6, 10, 12, 14, 18, 31, 38, 39) | Yes; No | Binary | Qualifier value | 385432009 |
| Respiratory distress (10, 12, 21, 38, 39) | Yes; No | Binary | Clinical finding | 386661006 |
| Myalgia (9, 12, 18, 38) | Yes; No | Binary | Clinical finding | 36523521 |
| Headache (10, 14, 18, 38, 39) | Yes; No | Binary | Clinical finding | 43724002 |
| Nausea/ vomiting (4, 9, 14, 18, 21, 36) | Yes; No | Binary | Clinical finding | 65124004 |
| GI symptoms (4, 10, 15, 16, 39, 40) | Yes; No | Binary | Clinical finding | 664563201 |
| Anosmia (12, 16, 21, 34) | Yes; No | Binary | Situation | 162298006 |
| Runny nose (12, 13, 15, 34, 39, 41) | Yes; No | Binary | Situation | 162062008 |
| Sore throat (4, 12, 13, 16, 21, 34, 40, 41) | Yes; No | Binary | Situation | 162104009 |
| Unexpected fatigue (12, 13, 15, 16, 40) | Yes; No | Binary | Clinical finding | 93559003 |
| Real-Time Vital Sign Monitoring | ||||
| Oxygen saturation (SO2) (13, 18, 34) | 75 < mmHg; 75 – 100 mmHg; 100 > mmHg | Forced choice | Clinical finding | 448225001 |
| Heart rate (bit per minute) (10, 12, 13, 18, 34, 36, 41) | < 60 bps; 60-100 bps; > 100 bps | Forced choice | Clinical finding | 76863003 |
| Blood pressure (mmHg) (10, 12, 14, 16, 31, 39) | < 120; 120-139; > 140 | Forced choice | Clinical finding | 2004005 |
| Body temperature (°C) (2, 3, 12-14, 16, 21) | < 37.3; 37.3 – 39; > 39.0 | Forced choice | Clinical finding | 50177009 |
| Respiratory rate (breaths per min) (2, 3, 12, 16, 21) | ≤ 24; > 24 | Forced choice | Clinical finding | 289100008 |
| Occupational Criteria | ||||
| Employment status (5, 7, 8, 18, 34, 41) | Unemployed; Employed | Forced choice | Clinical finding | 224363007 |
| Working status (7, 16) | Full time; Part time | Forced choice | Clinical finding | 160903007 |
| If employed, occupation risks (3, 8, 13, 31, 34, 38, 39) | High risk; Medium risk; Low risk | Forced choice | Event | 16090731000119102 |
| Work situation during general quarantine (7, 16, 34, 38) | Not working; Working at usual place; Teleworking; Other | Forced choice | Clinical finding | 302201002 |
| Work in a patient care setting (3-5, 7, 9, 13, 16, 21, 39-41) | Yes; No | Binary | Clinical finding | 302201002 |
| Attending work at the time of symptom occur (4, 9, 10, 14, 34, 40) | Yes; No | Binary | Clinical finding | 83408003 |
| Travel/Relocation Data | ||||
| Recent travel / relocation (4, 6, 8, 10, 13, 15, 18, 34, 36, 40) | Yes; No | Binary | Situation | 473087005 |
| Reason for travel (6, 9, 15, 36, 41) | Holiday business; Pilgrimage other | Forced choice | clinical finding | 161091009 |
| Travel type (4, 6, 8, 18) | Domestic travel; Foreign travel | Binary | Observable entity | 441969007 |
| Date of departure (3-6, 8, 9, 14, 34, 40) | dd/mm/yy | Integer | Observable entity | 810811000000107 |
| Number of travels in the last 7 days (5, 6, 8, 9, 11, 16, 36, 40) | None; One - two times a week; Two - four times a week; More than five times a week | Forced choice | Qualifier value | 259083004 |
| Travel to epidemic places (2, 5, 7, 9, 18, 21, 36, 40) | Yes; No | Binary | Clinical finding | 506931000000109 |
| Relocation / transfer method (3, 5, 9-11, 13, 16, 36) | Public transportation; Personal transportation | Binary | Procedure | 715957006 |
| Duration of travel (3, 7, 13) | Daily travel (1 day <); 1 day ≥ | Binary | Qualifier value | 69620002 |
| Contact Tracing Data | ||||
| Prior contact tracing experience (10, 15-18, 22, 25) | Yes; No | Binary | Procedure; | 225368008 |
| If yes, prior contact tracing approach (2, 3, 7, 13) | Conventional; Automatic | Binary | Clinical finding; | 52669001 |
| If Automatic, contact tracing technology (2, 3, 5-11, 14, 18, 31, 34, 40, 41) | Mobile phone; Implant tools other microcomputers | Forced choice | Qualifier value | 723991000000105 |
| Contact tracer ID (3, 5, 6, 9, 11, 14, 34, 38, 39) | XXXX | Forced choice | Qualifier value | 118522005 |
| Notification ID (3, 5, 6, 9, 10, 34, 39) | XXX /XXXX -X | Forced choice | Observable entity | 895571000000108 |
| Contact Data | ||||
| Contact type (4, 10, 12, 13, 18, 36, 38) | Primary: Person-to-person; Secondary: Person-to-surface / animal | Binary | Social concept | 70862002 |
| Contact category (2-4, 14, 21, 31) | No contact; Family members; Social contact; other | Forced choice | Clinical finding | 381441000000103 |
| Contact risk level (13, 15, 34, 36, 41) | Living with an infected/suspected case in the past 14 days; Prolonged direct contact in the past 14 days; Casual and indirect contact in the past 14 days; Not in contact | Forced choice | Situation | 76906009 |
| Contact with care facility | Yes; No | Binary | Situation | 136569214 |
| Contact frequency (3, 8, 16, 18) | Sometimes: ≥ 2 times a day; Always: 2 - 4 times a day; Repeatedly: < 4 times a day | Forced choice | Qualifier value | 735269004 |
| Contact list (person) (7, 13, 34) | 5 >; 5 - 10; 10 - 30; 30 < | Forced choice | Social context | 125676002 |
| Minimal distance of contact (meters) (2-4, 14, 21, 36, 41) | 2 >; 2 < | Binary | Qualifier value | 421669002 |
| Date of last contact (6, 12, 14, 16, 18, 21, 36, 41) | yyyy /mm/ dd | Integer | ||
| Time between contact and diagnosis (10, 15, 16, 18, 41) | Numerical | Qualifier value | 305698526 | |
| Total duration of contact (minutes) (3, 7, 8, 13, 34, 36) | ≥ 15; < 15 | Binary | Qualifier value | 356624006 |
| Total duration of contact (day) (3, 7, 8, 13, 34, 36) | ≥ 14; < 14 | Binary | Qualifier value | 258703001 |
5. Discussion
Contact tracing is known as a crucial surveillance measure in avoiding the spread of epidemic diseases such as the current COVID-19. During this epidemic, contact tracing data should be integrated across healthcare data collection systems at the national level (34). However, data are gathered from stand-alone recording and reporting systems largely manually generated via the contact tracing process. Data collection is a crucial strategic preparation measure for governments and health officials battling the COVID-19 epidemic (36).
The CoV-CT-MDS is a promising tool to meet some of the data necessary for epidemiology contact tracing leading to a validated template for the documentation of active case finding for public health practice and research purposes. Determining a core data set or MDS from a scientific perspective and according to the actual demands of users is the most central prerequisite for the design and development of any information system or app in the healthcare industry (38). It can be advantageous for designers and vendors of health information systems to simplify and accelerate the development of such systems and reduce the possibility of their failure (39). From this point of view, in this study, the CoV-CT-MDS can be used as a basis for the effective collection and management of data related to COVID-19 contact tracing using related information systems or apps.
In the initial months of the pandemic, contact tracing measures were recorded through manual data collection tools (eg, in Excel sheets, spreadsheet), which was a time-intensive, resource-demanding, and error-prone process (18, 40). Additionally, the conventional approaches did not always offer inclusive data about the number of investigated contacts, the nature of the relationship between cases and contacts, the number of contacts, who in turn, become cases, and the first and last days of follow-up surveillance (21, 41). To cope with these issues, it is essential to develop a contact tracing system that enables standardized data recording and accelerates the surveillance of contacts and outbreak paths (31, 42). This system allows intervallic analyses for the creation of standard reports and offers detailed epidemiological analysis for the identification of high-risk exposures and targeting of contact tracing efforts (21, 41, 43).
Implementing an active and responsive contact tracing strategy would be a valuable containing measure for avoiding the transmission of COVID-19. In this context, mobile technology enabling self-reports and smartphone applications for virtual contact tracing could be used to control disease outbreak and detect as well as quarantine COVID-19 cases and those who may have been exposed to the virus (44). For this purpose, a contact tracing system including timely and accurate data collection process and a unified case reporting template are proposed to guide healthcare authorities for proper interventions (20, 32, 36). There is, therefore, a pressing need for a unified data collection template to swiftly and prospectively collect high-quality data related to recent exposure and mobility patterns of confirmed and suspected individuals (45, 46).
The novelty of COVID-19 with frequent mutations of the virus demands numerous and unknown aspects to be investigated in prospective studies, and thus, studies related to COVID-19 contact tracing are limited at the time of writing this article (Decembers 2020). Hence, the main limitation of this living systematic review is the scarcity of available related resources and lack of data enrichment. Review of only English-language articles is another limitation of the study. However, multiple scientific databases were broadly reviewed. Future modifications, along with a Delphi survey is recommended to augment the COV-CT-MDS.
5.1. Conclusions
An effective COVID-19 contact tracing system requires reliable and timely information to guide fully informed decisions to contain the further spread of the disease by taking early preventive actions. For developing the CoV-CT-MDS, we performed an extensive literature review and expert view to identify the proposed contact tracing data fields and corresponding variables from an evidence-based perspective. The COV-CT-MDS as a unified data collection tool is the first step for developing a mobile-based contact tracing system. This template can provide valuable information for clinicians, health policymakers, and researchers for integrating the COVID-19 contact tracing efforts across Iran’s healthcare system. Given the prominence of reliable, accurate, and comprehensive data on COVID-19 surveillance measures, it is suggested that different countries design and implement a comprehensive national MDS for COVID-19 contact tracing.