1. Background
Cancer is one of the most life-threatening issues affecting human health worldwide. Colorectal cancer (CRC) is the third most common cancer in men and women in the United States (1). Graduate lifestyle changes (e.g., less smoking, aspirin use, etc.) by using monitoring tests and new preventive drugs have decreased the mortality and incidence rates of CRC. At least one-third of the CRC complications could be prevented by healthy behavior, normal body weight, and less smoking or alcohol use. Further studies are needed to clarify that the higher CRC burden in young adults causes and treats low response tumor subtypes (1). In Iran, CRC is the third common cause of malignancy, and its incidence rate has been raised due to an increase in individual and environmental risk factors, improvement in the registry system, and access to health services (2). CRC is found to be a multifactorial disease, meanwhile, the hereditary route of CRC occurrence is crucial, which showed 20 percent in this malignancy (2).
Viral infections as an extrinsic cause of cancer are an interesting topic, and the role of some viruses such as Epstein-Barr virus (EBV), hepatitis C virus (HCV), hepatitis B virus (HBV), human papillomavirus (HPV), human herpes virus 8 (HHV8), and human T-lymphotropic virus type 1 (HTLV-I) in cancer production are clarified to some extent (3).
HPVs are a major genus in the papillomaviridae family, consisting of above 120 species divided into high-risk (HR) and low-risk (LR) types. HPVs have been investigated for their role in different malignancies such as cervical, penile, vulvar, prostate, anal, ovarian, skin, and urinary tract. Recurrent respiratory papillomatosis (RRP), genital warts, and colorectal cancers are controversial issues associated with HPVs. Some studies on Iranian CRC patients reported the controversial rates of HPV infection and their relationships with CRC (4).
2. Objectives
In this regard, we aimed to investigate the association between HPV infection with CRC in Iranian patients using a molecular survey for the phylogenetic analysis and molecular determination of HPV-16 genome status.
3. Methods
3.1. Study Setting
The present case-control study was conducted by hospitals affiliated with the Iran University of Medical Sciences, Tehran, Iran. The participants were selected according to the diagnosis of sophisticated pathologists considering inclusion criteria.
The participants underwent total endoscopy and colonoscopy by a Fujinon machine (Fujinon, Japan) and were divided into two groups. The cases were diagnosed with CRC malignancy and were sporadic and non-familial cancerous. The controls were diagnosed with non-malignant lesions. All patients read and signed informed consent forms, and patient data were extracted from the medical record repository from June 2011 to December 2018. Archived formalin-fixed paraffin-embedded (FFPE) blocks were collected from patients, and their relevant hematoxylin and eosin (H&E) stained slides were reviewed by experts to select the correct blocks.
3.2. Nucleic Acid Extraction
The vibrating microtome (Leica VT 1000S, Leica Microsystems) was used for 20 microns dissected FFPE blocks and then deparaffinized using the Xylene method. QIAamp® DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) was performed for viral genome extraction according to the manufacturer’s instructions. NanoDrop ND-1000® (Thermo Fisher Scientific Inc., Waltham, MA, USA) spectrophotometry was used to evaluate isolated nucleic acid. Purified DNA was kept at -20°C until it was used.
3.3. HPV Detection
HPV DNA was detected by using the nested-PCR (nPCR) method. The universal primers MY09/11 (outer) and GP5+/6+ (inner) were used for L1 amplification and the detection of HPV-positive specimens according to previous protocols (5, 6). The Bio-Rad T100™ Thermal cycler was used for the polymerase chain reaction. Furthermore, Beta globin as an external PCR control was used following the previous protocols (5, 6). The heating program was as follows: 1st at 95°C for 5min, 45st at 95°C for 30 s, 56°C for 30 s (for outer primers), 48°C for 30 s (for inner primers), 72°C for 45 s, and 1st at 72°C for 5 min. The first-round PCR products were used as a template for the second-round PCR. The 25 µL reaction mixture consisted of 12.5 uL AMPLICON 2x PCR mix (AMPLICON co., Denmark), 1 µL each forward or reverse primers, and 10 µL extracted sample DNA (200 - 500 ng concentration), and distilled water was also added to reach the rest of the total of 25 uL reaction mixture volume.
3.4. HPV Typing
INNO-LiPA HPV genotyping extra assay (Innogenetics) was used for HPV typing to detect HR HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, 82; probable high risk types 26, 53, 66; LR HPVs 6, 11, 40, 43, 44, 54, 70; and unknown-risk types 69, 71, 74 according to previous protocols (7).
3.5. HPV DNA Physical Status
In this study, HPV-16 genome status was investigated using previous protocols (8, 9). Briefly, in the integrated form of HPV, some regions, such as E6, E7, LCR, and L1 were inserted into the host genome, and the other genes such as E2 were removed. We used the E2/E7 ratio to detect three integrated, episomal, and mixed forms in HPV-16 using the quantitative real-time PCR assay (qRT-PCR). Specific primers for E2 and E7 genes were obtained from Cheung et al.’s study (8, 9). Moreover, qRT-PCR was done using Rotor-Gene-Q 6000 thermocycler (Corbett, Australia) setting. The 15 µL reaction mixture included 7.5 µL of 2 × Amplicon III mix (Odense M, Denmark) and 0.5 µL of each primer, and distilled water was also used to reach the rest of the total 15 uL reaction mixture volume. The heating protocol for E7 was 1st at 95°C for 5 min, 40st at 95°C for 30 s, and 60°C for 30 s. Absorbance was adjusted to 60°C for each cycle. The heating protocol for E2 included 1st at 95°C for 5 min, 45st at 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. Absorbance was adjusted to 72°C for each cycle. Standard curves were drawn on the serial dilution of the HPV-16 E2 and E7 gene standards cloned into the PUC57 plasmid separately. A commercially available cloning system (TA Cloning kit of Invitrogen, San Diego, CA) was used for cloning according to the manufacturer’s protocol.
3.6. Nucleotide Sequencing and Phylogenetic Analysis
The PCR products of amplified HPV-positive subsets were purified using a High Pure PCR Product Purification Kit (Roche Diagnostic GmbH, Mannheim, Germany) according to the manufacturer’s instructions. Bidirectional sequencing was done by an ABI 3730 XL sequencer (ABI, Foster City, California). CLC Main Workbench 5 (CLC Bio, Aarhus, Denmark) and MEGA5 (www.megasoftware.net) software were used to compare and analyze the sequences. After analysis of the sequences via an online bioinformatics software the basic local alignment search tool (BLAST), the HPV reference sequences NC_001526.4 and NC_1596.1 were used for HPV-16 and HPV-9 phylogenetic tree, respectively.
3.7. Statistical Analysis
SPSS software version 22 (SPSS Inc., Chicago, IL, USA) was used to analyze the collected data. In descriptive statistics, frequencies were calculated and presented in a table. In analytical statistics, the chi-square test was performed to evaluate the association between categorical or nominal variables. In this study, P < 0.05 was set as the significance level.
4. Results
Of 157 participants, 66 individuals were CRC cases, and 91 subjects were healthy non-malignant controls. Mean age and standard deviations in the case and control groups were 59.35 ± 14.48 and 57.21 ± 14.66, respectively. Moreover, 57.6% of the cases and 57.1% of the controls were male.
According to the findings, a positive HPV DNA test was detected in 9.1% (6 patients) of cancerous patients as compared to 3.3 % (n = 3) of patients with colorectal polyps (P = 0.168) (Appendix 1). No significant relationship was noticed between the location of the tumor and the presence of HPV DNA in relevant tissues (P = 0.934).
Our findings revealed a significantly higher mean age (67.78 ± 10.08) in individuals with positive HPV DNA than in those with a negative HPV DNA (57.52 ± 14.63) (P = 0.040). Of nine HPV positive individuals and 148 HPV negative individuals, six (66.7%) and 84 (56.8%) patients were male, respectively (P = 0.559).
Of the nine HPV-positive individuals, the HPV genotyping of the samples of four patients could be determined by the PCR-sequencing method based on their higher viral load and purity, and only three HPV-16 isolates were confirmed by INNO-LiPA HPV genotyping. Using the PCR-sequencing method, we found four HPV genotypes, including HPV-16 (n = 3) and HPV-9 (n = 1). Another five HPV-positive isolates were detected based on gel electrophoresis and the band size from PCR products. The results of the INNO-LiPA HPV genotyping method confirmed our three HPV-16 results; however, no HPV-9 could be detected due to the limitations of the kit.
Of typeable samples (n = 4), two (out of 3 samples) were HPV-16, and one sample was HPV-9 in HPV-positive cancerous tissues (n = 6). In polyps (n = 3), only one sample was typeable and detected with HPV-16 infection. The non-typeable cases in the CRC and control groups were three and two cases, respectively. Statistically, there was no significant correlation in these groups (P = 0.75).
Table 1 shows the demographic and anatomical features of individuals with polyps based on the presence of HPV. No significant association was detected between the demographical and anatomical variables with the presence of HPV in individuals with polyps.
| Variables | HPV+ (n = 3) | HPV- (n = 88) | P-Value |
|---|---|---|---|
| Gender | 0.608 b | ||
| Male | 2 (66.7) | 50 (56.8) | |
| Female | 1 (33.3) | 38 (43.2) | |
| Age group (y) | 0.055 b | ||
| < 40 | 0 (0) | 14 (15.9) | |
| 40 - 64 | 0 (0) | 48 (54.5) | |
| ≥ 65 | 3 (100) | 26 (29.5) | |
| Tumor location | 0.879 b | ||
| Colon | 1 (33.3) | 28 (31.8) | |
| Rectum | 1 (33.3) | 24 (27.3) | |
| Cecum | 0 (0) | 16 (18.2) | |
| Ileum | 0 (0) | 5 (5.7) | |
| Sigmoid | 1 (33.3) | 15 (17) |
a Values are expressed as No. (%).
b Fisher’s exact test was used.
Table 2 presents the demographic, anatomical, and pathological features of individuals with colorectal malignancy based on the presence of HPV. No significant relationship was detected between these features and the presence of HPV in individuals with polyps.
| Variables | HPV+ (n=6) | HPV- (n=60) | P-Value |
|---|---|---|---|
| Gender | 0.696 | ||
| Male | 4 (66.7) | 34 (56.7) | |
| Female | 2 (33.3) | 26 (43.3) | |
| Age group (y) | 0.745 | ||
| < 40 | 0 (0) | 8 (13.3) | |
| 40 - 64 | 3 (50) | 29 (48.3) | |
| ≥ 65 | 3 (50) | 23 (38.3) | |
| Tumor location | 0.750 | ||
| Colon | 3 (50) | 26 (43.3) | |
| Rectum | 2 (33.3) | 11 (18.3) | |
| Cecum | 1 (16.7) | 10 (16.7) | |
| Ileum | 0 (0) | 2 (3.3) | |
| Sigmoid | 0 (0) | 11 (18.3) | |
| Tumor stage | 0.912 | ||
| T | 0 (0) | 1 (1.7) | |
| T1 | 0 (0) | 5 (8.3) | |
| T2 | 1 (16.7) | 8 (13.3) | |
| T3 | 3 (50) | 33 (55) | |
| T4 | 2 (33.3) | 13 (21.7) | |
| Mucinous tumor | 0.659 | ||
| Yes | 1 (16.7) | 19 (31.7) | |
| No | 5 (83.3) | 41 (68.3) | |
| Differentiation status | 0.408 | ||
| Well | 2 (33.3) | 34 (57.6) | |
| Moderate | 3 (50) | 18 (30.5) | |
| Poor | 0 (0) | 1 (1.7) | |
| Undifferentiated | 1 (16.7) | 6 (10.2) | |
| Tumor grade | 0.682 | ||
| High grade | 3 (50) | 24 (40) | |
| Low grade | 3 (50) | 36 (60) | |
| Lymph node involvement | 0.344 | ||
| Yes | 3 (50) | 16 (26.7) | |
| No | 3 (50) | 44 (73.3) | |
| No | 5 (83.3) | 55 (91.7) |
a Values are expressed as No. (%).
4.1. HPV-16 Physical Status
Each of the three HPV-16-positive strains underwent two qRT-PCR for the quantitation of E2/E7 genes (Appendix 2). The results showed an episomal form of HPV-16 in one of the women in the healthy control group, who had no malignant lesions. The integrated and mixed forms of HPV-16 were found in two males in the CRC cases.
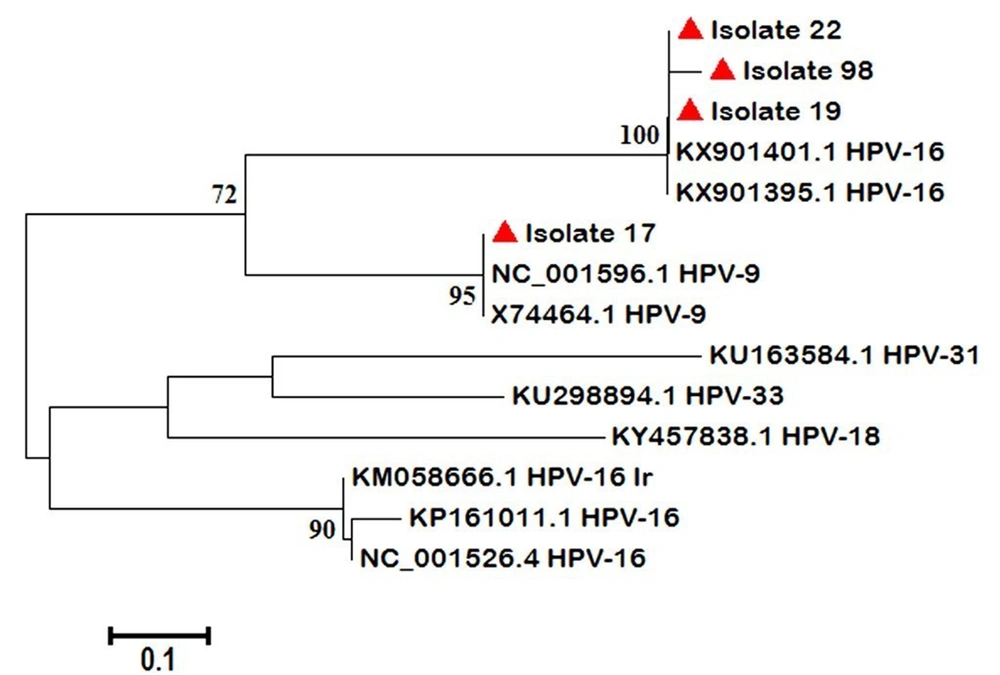
The phylogenetic tree was drawn by four HPV positive samples (three were from the case group (isolates 19, 22, and 17) and one from the control group (isolate 98) groups) (Figure 1).
5. Discussion
HPV as a worldwide-distributed viral infection with cancer as its likely outcome, especially in patients infected with HR HPV types, has been examined in different malignancies. The relationship between HPV and CRC progression has been investigated, and its potential role in CRC progression has been reported (10-12). Accordingly, based on the high rate of CRC, especially in elderly aged above 50 years, and its major complications in Iran, we aimed to investigate the role of HPV in these patients in a case-control study setting, and then we used them for the phylogenetic analysis and molecular determination of HPV-16 genome status. We analyzed 157 patients who underwent total colonoscopy and, regarding their complications, they were divided into two CRC (n = 66) and healthy (n = 91) groups. The HPV investigation was performed by the PCR-sequencing method, and the results were confirmed by INNO-LiPA HPV genotyping. Then a phylogenetic tree drawn on HPV positive isolates sequences and HPV-16 isolates physical genome status was determined by qRT-PCR for the first time in Iran. It was a rare report on CRC worldwide.
Our findings showed a similar rate of HPV infection in the CRC patients and healthy controls, and there was a significant correlation between HPV infection and higher ages (P = 0.04). Furthermore, the HPV-16 genotype was prevalent in our studied population. Although most studies using conventional and nested PCR methods reported low rates of HPV infection, other techniques such as immunocytochemistry techniques have reported higher rates. It seems that more accurate methods and confirmatory tests should be used to reach more comprehensive results.
Iran, as a WHO Eastern Mediterranean region and a middle-East country, had a rich background of HPV infection in different cancers, including cervical (77.5 %), head and neck (32.4%), esophageal squamous cell (23.1%), lung (22.2%), prostate (10.4%), urinary tract (30.9%), breast (14%), and skin (25.2%) cancers. HPV16 and 18 were the most frequent HPV types in all HPV-related cancers (13). Recently, colorectal cancer as a major malignancy with an increasing rate of complications, incidence, and mortality, has been considered in HPV investigations. Some studies in different provinces of Iran have reported HPV involvement in CRC patients to range from 0 to 35%. For example, Nosrati et al. (14) in Northern Iran studied 95 CRC patients and 95 healthy colon tissue using the conventional-PCR (cPCR) method; Aghakhani et al. (15) in Tehran, the capital of Iran, examined 70 colon adenocarcinoma and 30 adenoma cases using the nPCR technique. Taherian et al. (16) in Tehran, the capital of Iran, studied 50 CRC and 50 non-malignant colon tissues using the cPCR technique. These researchers found no HPV-positive results. On the other hand, Meshkat et al. (17) in Mashhad, northeast of Iran, showed that one (1%) out of 100 CRC patients was HPV positive using the cPCR technique; Mahmoudvand et al., (4) in Shiraz, southwest of Iran, reported that HPV DNA was noticed in 2.8% (6/210) of the samples, including 2.8% (2/70) adenocarcinoma and 5.7% (4/70) adenomatous polyps, using the cPCR technique. Their study showed no positive result in 70 healthy colon tissues. HPV-16 was the most prevalent (83.3%), followed by HPV-18 (66.6%), and four samples were co-infected by both genotypes. Ranjbar et al. (18) in Tehran, the capital of Iran, reported HPV DNA in 6.2% (5/80) of CRC cases and 1.2% (1/80) of healthy colon tissues using the nPCR technique. In Malekpour Afshar et al.’s (3) study in Kerman, southeast Iran, 22.6% (19/84) of the cases were positive for HPV infection in CRC patients, and HPV-16 frequency was reported in 10.6% of the cases using the qRT-PCR SYBR Green method and the INNO-LiPA HPV genotyping assay. Motlagh et al. (19) in Tehran, the capital of Iran, showed that 35% (21/60) of tissue sections from patients with CRC were HPV DNAs positive using immunocytochemistry techniques, of whom 32% (19/60) were HPV-18, and 18% (11/60) were HPV-16.
Interestingly, regarding a neighbor-joining phylogenetic tree, our HPV-16 positive strains had close similarity to Uruguayan HPV-16 strains and were categorized in different branches from previous Iranian records (20)
HPV-related carcinogenesis triggered by chromosomal instability and an integrated HPV genome could establish a more severe disease, and then the viral load gradually decreased. In reported cancerous cells, the L1, E6, E7, and LCR regions were integrated into the host genome (8, 21-24). The integration of viral DNA causes the overexpression of some viral oncoproteins affecting host proto-oncoproteins such as pRB and p53 (25, 26). The HPV genome status can be used as a prognostic marker in HPV-associated cancers (25, 27-29).
The integration of the HPV genome in different cancers and malignancy stages has been reported. Bernabe-Dones et al. studied 12 HPV-16 infected CRC patients to evaluate HPV genome status using nPCR for E2 gene. They found an integrated form in all patients. Accordingly, these researchers concluded that the integration of the HPV genome could be associated with colorectal carcinogenesis (11). The findings showed that the HPV-16 positive CRC group was integrated and mixed forms, and there was an episomal form in the control group. The mixed form consisted of a mixture of integrated and episomal forms. From this perspective, the findings of the present study are consistent with their findings. According to the histological and pathological data, the episomal form of HPV-16 positive was in a polypoid lesion colon, which could have an impact on the higher risk of establishing a malignancy. However, we found a few cases to conclude. The initial stages of chromosomal instability and consequent neoplasia could involve an episomal form of the HPV infection. Although our CRC patients were involved in high-grade cancerous tissue that the malignancy development could accelerate by HPV integration, our limited positive cases restricted making a comprehensive conclusion.
The small sample size and the use of one location of cancerous tissues to detect viral infection were the limitations of the present study. Accordingly, we suggest an extra marginal section for the cancerous population and the other tests except for PCR-based ones for HPV detection.
In conclusion, the present study showed a positive HPV DNA test in 9.1% of the CRC cases and revealed a correlation between HPV infection and older ages. A majority of the detected HPV genotypes were HPV-16 in the CRC patients; however, there was no significant relationship in our limited positive cases. There is a remarkable rate of HPV infection in the CRC patients and healthy colon tissues. Further studies are recommended to include larger sample sizes and use different accurate confirmatory tests to reach more comprehensive results.