1. Background
Coronavirus Disease 2019 (COVID-19), a viral infectious disease, generally causes severe respiratory syndrome and gastrointestinal and central nervous system complications in patients infected by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1). The COVID-19 pandemic has posed a severe global burden, claiming more than 0.8 million lives throughout the world between December 2019 and August 2020 (2, 3). Cytokine storms in COVID-19 patients can cause intensive inflammatory events and a highly pro-thrombotic environment, leading to thrombotic complications (4). Thromboembolic complications are considered life-threatening in COVID-19 patients (5).
The prothrombotic state observed in COVID-19 patients is multifactorial, and platelets play an indispensable role in initiating the thrombotic process. Therefore, platelet activities and functions serve as potential hematologic parameters for monitoring thrombotic complications in COVID-19 patients (6, 7). Studies have shown that platelets in COVID-19 patients secrete elevated levels of IL-1beta, soluble CD40L (8), serum circulating serotonin, and PF4 compared to healthy controls. Moreover, platelet activation cell markers, including P-selectin and CD63, are increased in critically ill COVID-19 patients, indicating a hyperactive phenotype of platelets (5).
Platelets play a crucial role in inflammation and immunity by modulating various leukocyte functions through the release of inflammatory mediators as well as direct cell-cell interaction. Previous studies have found that levels of circulating platelet-leukocyte aggregates increase in patients with inflammatory or thrombotic conditions (9).
Platelet-leukocyte interactions lead to leukocyte mobilization to the inflammation zone, the release of pro-inflammatory mediators, and neutrophil extracellular traps (NET) by leukocytes, oxidative burst, and phagocytosis (10-14). The platelet-leukocyte aggregates are associated with various thrombotic and inflammatory conditions, including severe lung injuries, rheumatoid arthritis, inflammatory bowel disease, and cerebrovascular and cardiovascular disease (15-19). The most important receptors involved in platelet-leukocyte aggregates include P-selectin (CD-62p), CD41/CD61 complex (Glycoprotein IIb/IIIa), and C40L on the platelets, and PSGL-1, Mac-1, and CD40 on the leukocytes (20-22). Monocytes have been shown to have a higher affinity for platelet complexes than other leukocytes, and these complexes serve as a more sensitive marker for platelet activation than P-selectin (23). The interaction between monocytes and platelets among leukocytes represents the most significant binding affinity between P-selectin and PSGL-1 (24-26).
Platelet activation and platelet-leukocyte interaction have been established in the pathophysiology of various viral infections, such as the dengue virus, human immunodeficiency virus (HIV), and influenza virus (27-34). Recent studies have also reported that COVID-19 patients have a higher level of activated platelets and increased interaction of platelets with neutrophils, monocytes, and T cells (35). Moreover, platelet-monocyte aggregate formation results in the expression of tissue factor in monocytes via P-selectin/αIIb/β3 integrin-mediated signaling. The binding of tissue factor with FVII proceeds coagulation cascade activation (5).
Interferon-induced transmembrane protein 3 (IFITM3) is a small protein that plays crucial roles against viral infection in both acquired and innate immune responses (36-45). Interferon-induced transmembrane protein 3 protein is commonly expressed on endosomes and lysosomes and eliminates the fusion of the viral membrane with the host cell membrane in a wide range of capsid viruses like influenza, Ebola, and Marburg (46). Moreover, IFITM3 is active against vesicular stomatitis virus, HIV, respiratory syncytial virus, and Zika virus (47, 48). In addition, IFITM3 is involved in several cellular processes like differentiation, apoptosis, cell adhesion, and modulation of immune cells which can boost immunity against influenza A, West Nile, and dengue viruses, and SARS-CoV (36, 39, 49, 50).
Given the significant social and economic burden of COVID-19, along with the important roles of platelet-monocyte aggregates in initiating inflammation and thrombosis, as well as the pivotal role of IFITM3 in virus elimination, this study sought to explore the association of platelet-monocyte aggregates and IFITM3 mRNA levels with coagulation tests and disease progression in patients with severe and mild/moderate COVID-19.
2. Objectives
By investigating the relationship between these biomarkers and COVID-19 severity, we hope to gain a better understanding of the underlying mechanisms of the disease and identify potential targets for future treatments. This study may also help to identify patients who are at higher risk for thrombotic complications and guide clinical decision-making.
3. Methods
3.1. Study Setting and Subjects
This cross-sectional study was conducted on 54 COVID-19 patients admitted to Ali-Asghar Hospital, Shiraz, Iran, from 2020 to 2021. The sample size was calculated according to the formula n = (Z1-α/2+Z1-β)2/cr2+3. The inclusion criteria were a positive PCR test for SARS-CoV-2, age over 18, and hospitalization time of less than 72 hours before sampling. Patients with a history of comorbidities, including cardiovascular disease, previous respiratory disease, cancer, infection with other acute or chronic viral infections like HIV, HCV, and influenza, were excluded from our study. Moreover, patients who were pregnant or under previous treatment with anticoagulants and antiplatelet regimens were excluded. The included patients were divided into two subgroups: Severe and mild/moderate, according to the WHO classification of COVID-19 severity based on SpO2 (mild/moderate > 94%, severe < 94%). Written informed consent was obtained from all eligible patients before their inclusion in the study.
3.2. Assessment of Medical Records of the Patients
Due to incomplete clinical files, some patients had to be excluded, leaving a final count of 54 patients. The general characteristics of the patients, including age, gender, and hospitalization period, were extracted from their medical records. Moreover, clinical tests including PT, PTT, INR, CBC indices (WBC, RBC, Hg, Lymphocyte, Neutrophil, and platelet count), O2 saturation, LDH, and C-reactive protein (CRP) were obtained from the medical records of the patients.
3.3. Sample Preparation
Four milliliters of venous blood samples were collected in two tubes, 2 ml each, with K2EDTA for each patient according to standard protocol. One series of tubes was assigned for flow cytometry. Another series was centrifuged at 3000 rpm for 7 minutes, and buffy coats were obtained in 1.5 mL micro-tubes and stored at -20°C until subsequent use.
3.4. Quantitative Real-time RT-PCR (q-RT-PCR)
To evaluate the mRNA expression level of the IFITM3 gene in the patients, q-RT-PCR was performed. First, RBCs in buffy coat samples were removed by adding RBC lysing solution and then washed with PBS three times. Then, the total RNA of buffy coat samples was isolated using Trizol (Invitrogen) according to the manufacturer’s protocol. The quality and quantity of extracted RNA were evaluated using Nanodrop (Thermo ScientificTM) and agarose gel electrophoresis. Next, single-strand cDNA synthesis was conducted using Easy cDNA Synthesis Kit (ParstousTM, Iran) according to the manufacturer’s instructions. To check the accuracy of the cDNA synthesis reaction, qPCR with a housekeeping gene primer, GAPDH, was carried out for all synthesized cDNA samples (the sequence and product size of the primers are shown in Table 1). After that, q-PCR was performed using the Cycler Rotor Gene Q instrument (Qiagen, Germany) with SYBR Green Master Mix (ParstousTM, Iran). The reaction contained SYBR Green (10 µL), cDNA (1 µL), forward and reverse primer (20 pmol), and nuclease-free water to a final volume of 20 µL. The thermocycling program included an initial heat activation and denaturation at 94°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, an annealing step for 30 seconds at 60°C, and an extension step for 30 seconds at 72°C. The mRNA expression level of IFITM3 was normalized with the GAPDH mRNA expression level. Melting curve and amplification plot analysis were performed to check qPCR reaction specificity. Moreover, the PCR products were electrophoresed on 2% agarose gel to evaluate the primers’ specificity. The relative expression level was calculated using the 2-∆Ct method as explained elsewhere. The reactions were conducted with at least two replicates and in the presence of no template control and reference gene in each experiment. All qPCR experiments were conducted according to standard protocol.
| Genes | Sequences (5’ to 3’) | PCR Product Size (bp) |
|---|---|---|
| IFITM3 | 173 | |
| Forward | ATAGCATTCGCCTACTCCGTG | |
| Reverse | CCATAGGCCTGGAAGATCAGCA | |
| GAPDH | 110 | |
| Forward | CACCAGGGCTGCTTTTAACTCTGGA | |
| Reverse | CCTTGACGGTGCCATGGAATTTGC |
The Sequence and Product Size of the Primers
3.5. Evaluation of Platelet-Monocyte Aggregates by Flow Cytometry
First, 50 μL of whole blood of COVID-19 patients were incubated with FITC-labeled anti-CD14 (Clone: HCD14, BioLegend) along with a PE-labeled anti-CD61 antibody (Clone: VIPL2, BD Biosciences, CA, USA) for 20 minutes in the dark at room temperature. Then, red blood cells were lysed with 1x ammonium chloride. Finally, samples were analyzed on BD FACSCalibur™ flow cytometer (BD Biosciences, CA, USA). The detection results were plotted on a CD14 vs. SSC dot plot for mononuclear cells (MNCs). Then, the CD14+ cells were plotted on a CD14 vs. CD61 dot plot. The CD14+CD61+ complexes were considered platelet-monocyte aggregates. Finally, data were processed using FlowJo TM v10 software.
3.6. Statistical Analysis
The data were analyzed using GraphPad Prism 9. The Shapiro test was used to check the normal distribution of the data. To determine the difference in the mean of the data, t-tests or Mann-Whitney tests were used for parametric and non-parametric data, respectively. The data were considered significant when the P-value was below 0.05. The correlation of variables was evaluated using the Spearman test. The r-value of 1 and -1 was considered a strong positive and reverse correlation, respectively.
4. Results
4.1. Patient Characteristics
This cross-sectional study included 54 hospitalized COVID-19 patients. According to the WHO classification for COVID-19 severity, 38.9% (n = 21) of the patients were classified as severe, and 61.1% (n = 33) as mild/moderate. Among those with severe COVID-19, 11 patients were male and 10 were female, while those with mild/moderate form comprised 15 males and 18 females. Furthermore, the majority (88.9%, n = 48) of the patients were in non-ICU wards, but six patients were in the ICU. Two subjects with severe COVID-19 expired during the study period. The overall average age of the patients was 49 ± 16.3 years, with a higher mean age observed in those with severe form compared to mild/moderate (52 ± 14.5 vs. 41 ± 17.38, respectively).
As expected, the lymphocyte count was significantly higher in the mild/moderate group (21.7 ± 8.9 vs. 16.3 ± 10.9, P = 0.02), while the neutrophil count was significantly higher in severe patients (78.3 ± 12.2 vs. 72.3 ± 9.9, P = 0.01). Furthermore, the levels of CRP and LDH were significantly higher in severe COVID-19 patients (P = 0.01 and P = 0.001, respectively). O2 saturation level was significantly higher in the mild/moderate group compared to severe patients (94.8 ± 0.9 vs. 85.4 ± 11.6, P < 0.001) (Table 2).
| Parameter | Severe (n = 21) | Mild/Moderate (n = 33) | Total (n = 54) | P-Value | |||
|---|---|---|---|---|---|---|---|
| Mean (± SD) | Min-Max | Mean (± SD) | Min-Max | Mean (± SD) | Min-Max | ||
| Age | 52 ± 14.5 | 32-86 | 47 ± 17.38 | 21 - 84 | 49 ± 16.3 | 23 – 86 | 0.28 |
| HP (days) | 10.48 ± 10.1 | 4-36 | 4.9 ± 1.0 | 3 - 7 | 7 ± 6.8 | 3 - 36 | 0.17 |
| O2 saturation | 85.4 ± 11.6 | 51-93 | 94.8 ± 0.9 | 93 - 97 | 91.1 ± 8.5 | 51 - 97 | < 0.001 |
| WBC (count × 103) | 6.6 ± 3.0 | 3.1-15.4 | 5.3 ± 1.7 | 2.5 - 9 | 5.8 ± 2.3 | 15.4 - 2.50 | 0.16 |
| Lymphocyte | 16.3 ± 10.9 | 5.6 - 38.6 | 21.7 ± 8.9 | 7.5 - 39.9 | 19.6 ± 10.0 | 5.6 - 39.9 | 0.02 |
| Neutrophil | 78.3 ± 12.2 | 53.6 - 92.2 | 72.3 ± 9.9 | 52.2-84.3 | 74.7 ± 11.1 | 52.2 - 92.2 | 0.01 |
| RBC (count × 106) | 4.8 ± 0.7 | 2.8 - 6.3 | 4.7 ± 0.5 | 3.7-5.9 | 4.8 ± 0.6 | 2.8 - 6.31 | 0.93 |
| Hg (mg/dL) | 13.7 ± 2.1 | 8.5 - 17.3 | 14.0 ± 1.6 | 10.9-19.7 | 13.9 ± 1.8 | 8.5 - 19.7 | 0.82 |
| PLT count (count × 103) | 182 ± 70 | 51 - 360 | 184 ± 60 | 75-329 | 183 ± 63 | 51 - 360 | 0.91 |
| PT (second) | 15.2 ± 2.8 | 11.5 - 22.9 | 15 ± 1.5 | 13-18 | 15.1 ± 2 | 11.5 - 22.9 | 0.76 |
| INR (IU) | 1.3 ± 0.3 | 0.8 - 2.3 | 1.2 ± 0.2 | 1-1.6 | 1.2 ± 0.2 | 0.8 - 2.3 | 0.82 |
| PTT (second) | 35 ± 8.9 | 22 - 65 | 37.3 ± 17.1 | 25-120 | 36.4 ± 14.5 | 22 - 120 | 0.9 |
| CRP | 58.8 ± 32.9 | 3 - 90 | 36 ± 28.2 | 3-88 | 44.9 ± 31.9 | 3 - 90 | 0.01 |
| LDH | 633 ± 157 | 279 - 859 | 498 ± 158 | 228-987 | 549 ± 169 | 228 - 987 | 0.001 |
Patients’ Characteristics and Their General Laboratory Findings
4.2. Interferon-Induced Transmembrane Protein 3 (mRNA Expression)
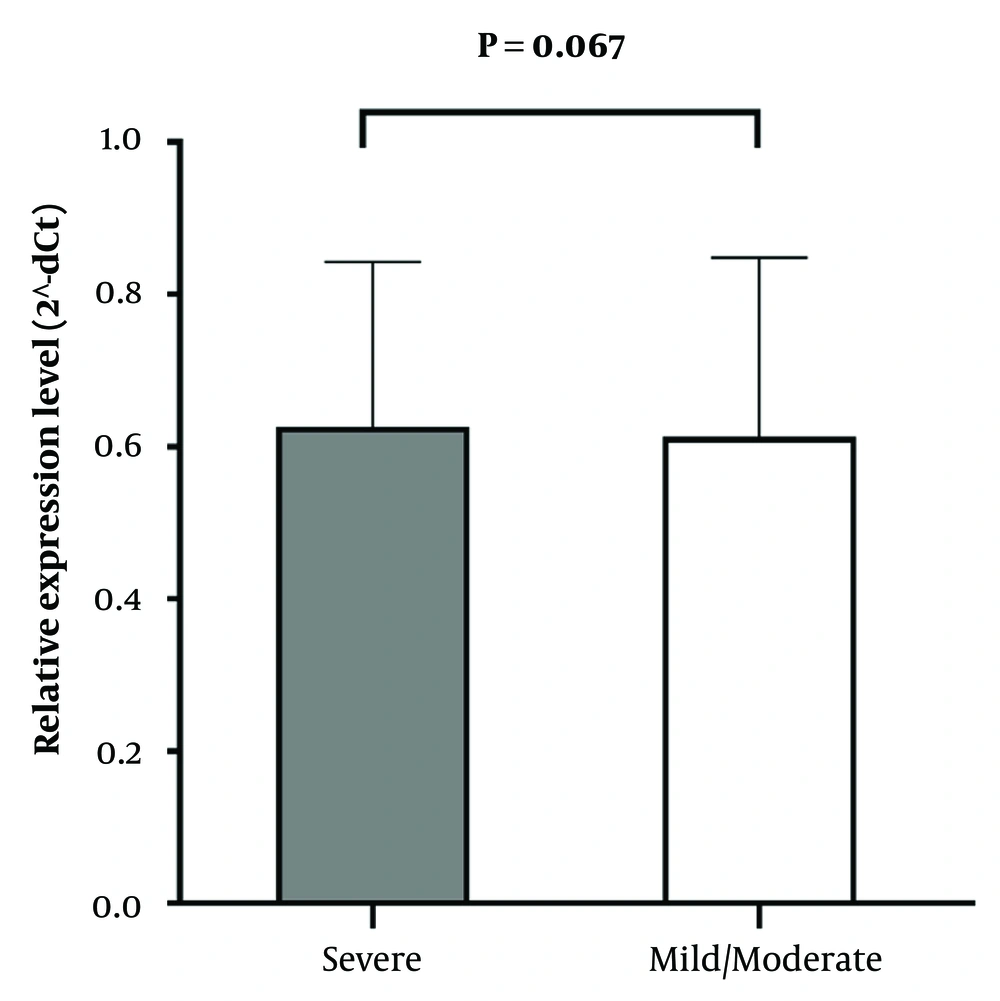
To evaluate the mRNA expression level of the IFITM3 gene, qRT-PCR was carried out. As shown in Figure 1, no significant difference was observed in the expression of the IFITM3 gene between severe and mild/moderate COVID-19 patients (P = 0.067).
4.3. Platelet-Monocyte Aggregate Formation Level
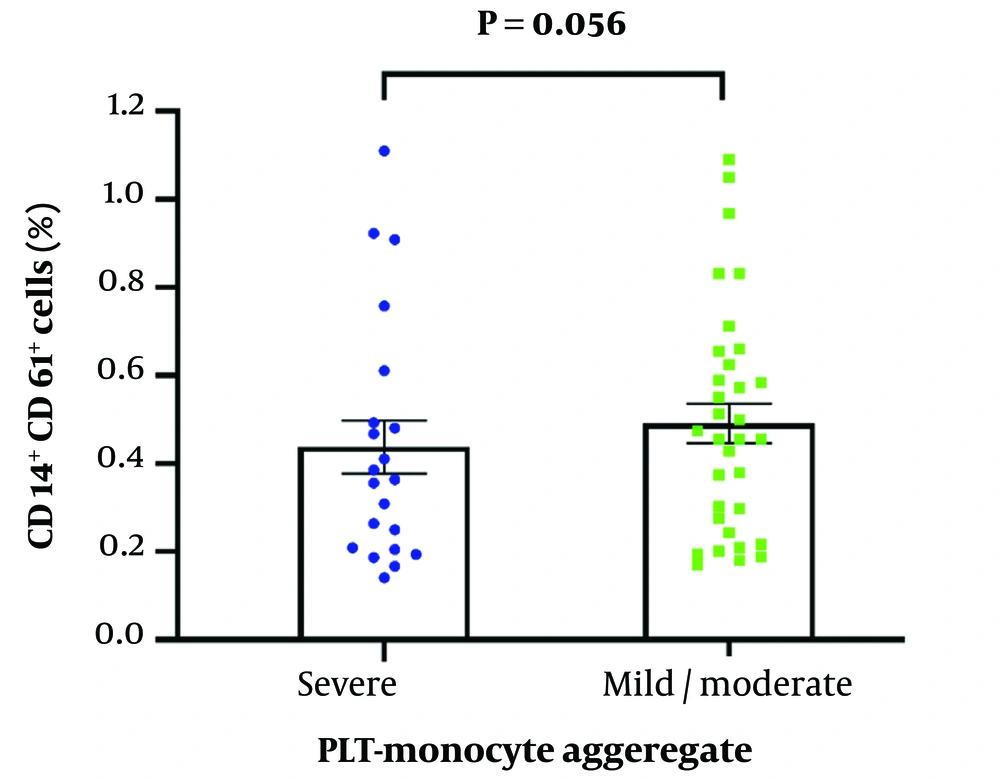
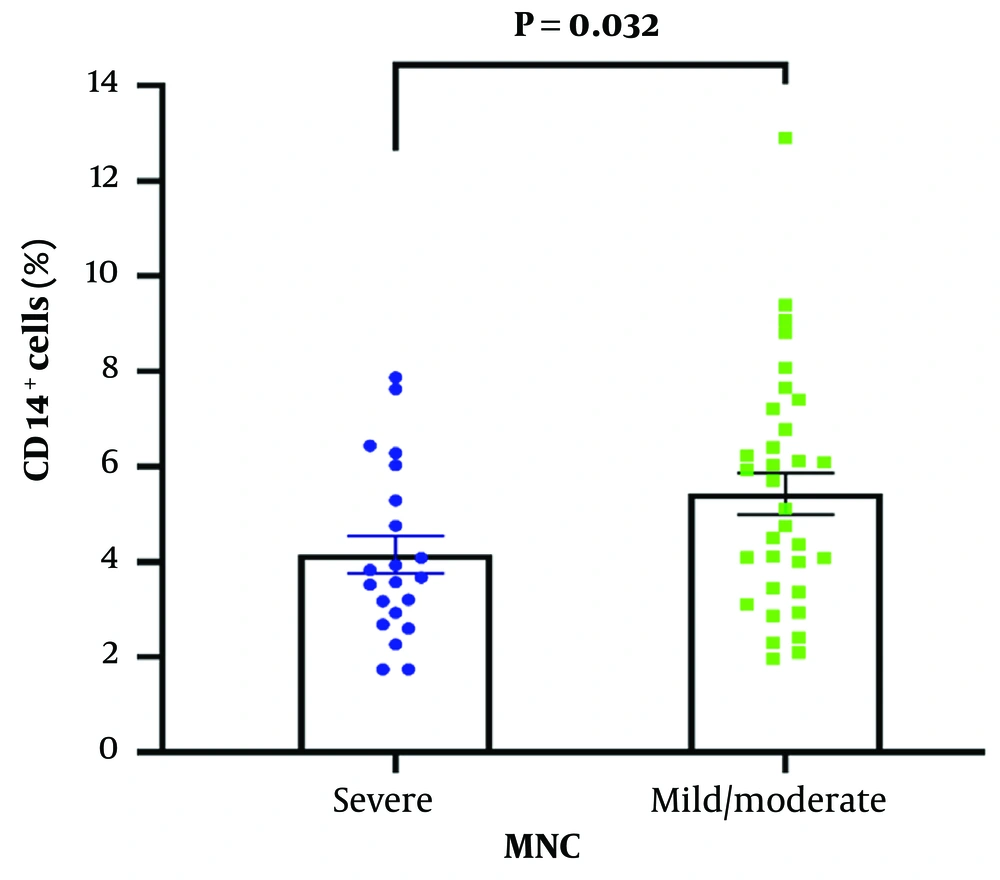
A flow cytometry assay was carried out to detect CD61 and CD14 cell surface markers to measure the level of platelet-monocyte complex formation. Although the percentage of MNCs was significantly higher in mild/moderate COVID-19 patients compared to severe patients (P = 0.032), there was no significant difference in platelet-monocyte complex formation between the two study groups (P = 0.056) (Figures 2 and 3). The key receptors contributing to platelet-leukocyte aggregates encompass P-selectin (CD-62p), CD41/CD61 complex (Glycoprotein IIb/IIIa), and CD40L on platelets, as well as PSGL-1, Mac-1, and CD40 on leukocytes (20-22). Monocytes exhibit a greater affinity for platelet complexes compared to other leukocytes, making these complexes a more sensitive marker for platelet activation than P-selectin (23).
4.4. Correlation of Variables
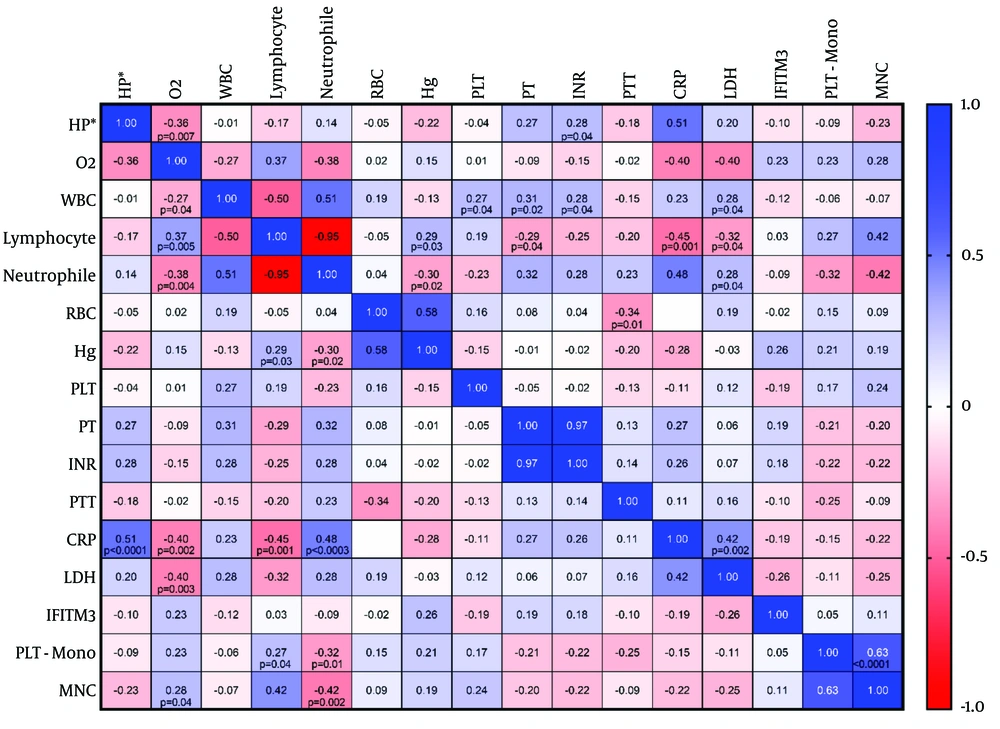
The Spearman statistical test showed that lymphocyte and MNC counts were positively correlated with platelet-monocyte aggregate formation (P = 0.04, and P < 0.001, respectively), while the neutrophil count was inversely correlated with platelet-monocyte aggregate formation (P = 0.01, r = -0.32) (Figure 4). However, the IFITM3 mRNA level was not correlated with any parameter. A strong positive correlation was observed between the hospitalization period and CRP (P < 0.001), CRP with neutrophils and LDH (P = 0.0003 and P = 0.002, respectively), as well as between O2 saturation and lymphocytes (P = 0.005). The correlation values of other variables are shown in Figure 4 as well.
5. Discussion
In this study, we sought to investigate the potential correlation between IFITM3 mRNA expression levels and platelet-monocyte complex formation with COVID-19 severity, as well as various inflammatory and coagulation markers, in a cohort of 54 hospitalized COVID-19 patients. We aimed to gain insight into the underlying mechanisms of COVID-19 pathogenesis and identify potential targets for therapeutic interventions.
We determined the mRNA expression level of IFITM3 in the buffy coat of COVID-19 patients using qRT-PCR and found no significant difference between our two study groups. Previous studies have reported that IFITMs (1, 2, and 3) can eliminate viral pathogens (36-50). However, the pro- and anti-viral activities of IFITMs have been reported for coronaviruses. A toxicogenomic analysis reported that IFITM3 is upregulated in SARS-CoV-2-infected human bronchial epithelium (51).
Interferon-induced transmembrane proteins have been found to hinder the entry of human coronaviruses, including SARS-CoV-1, SARS-CoV-2, and MERS-CoV, in artificial experimental conditions where viral particles were pseudotyped with Spike (S) proteins and cell lines overexpressed IFITMs (39, 52-56). However, a recent study found that endogenous IFITM expression assists SARS-CoV-2 membrane fusion with host human primary lung cells. The study showed that genuine SARS-CoV-2 Spike proteins hijack IFITMs to promote efficient viral infection and suggested that targeting IFITMs could inhibit SARS-CoV-2 infection of human lung cells (55). The reason behind the contradictory effects of overexpressed and endogenous IFITMs is not yet completely understood. However, it has been reported that specific mutations in IFITM3 affecting its topology may change its inhibitory role to an enhancer of coronavirus infection (52, 57). Moreover, two SNPs of IFITM3 have been associated with the risk of acquiring SARS-CoV-2 infection (56).
The small population of the present study may prevent the finding of the possible relationship between IFITM3 gene expression level and the prognosis of COVID-19 patients. Another point to consider is that most of the mentioned studies have examined the role of IFITM3 protein in the resistance to virus entry in lung epithelial cells, but our study examined the expression of this gene in blood monocytes. It is important to consider, however, that our study had the advantage of involving human subjects rather than an in-vitro experiment. Further studies with larger pre-genotyped study subjects may shed light on the mechanisms behind IFITM3.
We used flow cytometry to detect CD14, a well-known monocyte marker, and CD61 (GPIIIa), a platelet/megakaryocyte marker, to evaluate the level of platelet-monocyte complex formation. Our findings showed that the level of PLT-Monocyte aggregates was not statistically different between our study groups.
Activated platelets are connected with monocytes in PLT-Monocyte aggregates via P-selectin (58). We know that monocytes have fundamental roles in thromboinflammatory disease progression by their pro-inflammatory and coagulation roles. Increased levels of PLT-Monocyte aggregates are documented in many inflammatory and infectious diseases (58). There are well-established studies reporting increased levels of PLT-Monocyte aggregates in COVID-19 with positive correlations with more severe outcomes and thrombotic events (59-62). Inconsistent results, which show no or negative correlation of PLT-Monocyte aggregates with disease severity, have also been documented (63, 64). One obstacle of our study is the absence of a normal control group. Normal subjects were included in most of the mentioned studies that report increased levels of PLT-Monocyte aggregates.
These controversial results may be attributed to differences in the virus species of SARS-CoV infecting the patients, the time of blood sampling, and the consumption of platelet inhibitors like aspirin. Additionally, platelet count may be a confounding factor for the level of platelet-monocyte aggregates.
As expected, lymphocytopenia was significantly higher in severe COVID-19 patients compared to mild/moderate patients. Furthermore, significantly higher levels of LDH and CRP in severe patients indicate a more intense inflammatory environment. These results are consistent with previous studies on COVID-19 patients (65) and support the findings of Xiang et al., who reported severe lymphocytopenia, inflammatory cytokine escalation, and elevated CRP and LDH in fatal COVID-19 patients (66). Xiang et al. also observed severe tissue damage, lymphocyte apoptosis, and SARS-CoV-2 RNA accumulation in the spleen and hilar lymph node. However, our findings for coagulation tests, including PT and PTT, were not statistically different between our two study populations, which is consistent with a previous investigation (67).
We also observed a relatively strong positive correlation between the hospitalization period and CRP, a well-known inflammation biomarker, as well as between CRP and neutrophil count and LDH. In contrast, we observed a strong reverse correlation between the hospitalization period and O2 saturation, O2 saturation and neutrophil count, and CRP and LDH. Furthermore, we found a negative correlation between lymphocyte count and CRP and LDH. These findings are consistent with previous studies (68, 69) and suggest that a more inflammatory environment is associated with more severe clinical outcomes in COVID-19 patients.
5.1. Conclusions
Overall, the present study did not find any significant correlations between IFITM3 gene expression and platelet-monocyte aggregate levels with disease outcomes in COVID-19 patients. The research aimed to investigate the relationship between these biomarkers and COVID-19 severity to gain a better understanding of the disease mechanisms and potential treatment targets. While previous studies have shown the role of IFITM3 in eliminating viral infections, the results of this study did not show a clear association with COVID-19 severity. Monitoring inflammatory and coagulation markers remains crucial for managing COVID-19 patients. Further investigations with larger sample sizes as well as the investigation of aggregates with other sensitive methods may provide more insights into the mechanisms involved.