1. Background
Technological innovations in medical sciences like microarray and proteomics produce high throughput data sets, known as big or high dimensional data. In high dimensional data sets, the number of variables is very large whereas, and due to excessive costs, the sample size is typically small (1). Several studies have referred to challenges occurring in high dimensional settings as “curse of dimensionality” (2). The course of dimensionality leads to ill posed situation, when traditional statistical modeling is used. Traditional models are also inapplicable when the number of variables is greater than the sample size (3). To avoid the aforementioned challenges, researchers proposed penalized regression models and machine learning techniques in the last decades (4, 5).
Immunoglobulin A nephropathy (IgAN) is considered a chronic renal disease and the most prevalent glomerulonephritis throughout the world (6-8). The nature of diverse clinical presentations and prognosis in IgAN leads to poor prognosis as progress to end stage renal disease occurs nearly in 20% to 30% of patients and renal survival rate of half of them is less than 30 years (5, 9). Therefore, pathogenesis insight could contribute to early and non-invasive diagnosis of patients and increase survival and quality of life in IgAN patients. In the last decades, several studies have attempted to reach a safe diagnosis by investigating among IgAN biomarkers (10).
In order to model a large number of extracted biomarkers and identify the most effective biomarkers on IgAN disease, the researchers implemented 2 methods of penalized regression, known as LASSO and MCP, which are appropriate for high dimensional and low sample size settings. As an alternative method, variable selection using random forest method was used as a machine learning technique. Finally, the accuracy of all the 3 proposed models was compared and the panel of selected biomarkers and biological relevance were discussed.
2. Methods
In the current study, the data were obtained from the experiment of Samavat et al. on urinary protein profile of patients with IgA nephropathy and healthy individuals using nanoscale liquid chromatography with tandem mass spectrometry (nLC-MS system) (11). The case group in the mentioned study consisted of 13 patients with approved IgA nephropathy disease by biopsy and 8 healthy volunteers without any nephropathy disease, considered as the control group. Urinary protein profiles for both groups composed of 493 proteins. Therefore, the researchers encountered a high dimensional and low sample size problem and tried to implement appropriate statistical methods to handle this challenge.
2.1. Least Absolute Shrinkage and Selection Operator Logistic Regression
Least Absolute Shrinkage and Selection Operator (LASSO) is one of the simplest and well-known penalized methods that perform estimation and variable selection tasks, simultaneously. The amounts of shrinkage in LASSO are managed by a positive constant, known as tuning parameter. If be xi = (1, xi1, …, xip)T ,1 ≤ i ≤ n and β = (β0, β1, …, βp)T a p-dimensional vector of regression coefficients, LASSO logistic regression is defined as follows:
Equation 1.



and λ is the tuning parameter (12). Tuning parameter has a key role in all penalized methods and is estimated by k-fold cross validation (4, 12). In this study, the researchers used 5-fold cross-validation for estimating the optimal lambda.
2.2. Minimax Concave Penalty Logistic Regression
Minimax concave penalty (MCP), as a modern variable selection method, was introduced in 2010 by Zhang. Type of penalty in MCP leads to oracle property, which means that MCP is capable to estimate coefficients of all zero variables equal to zero and estimate coefficients of all non-zero variables as non-zero with a probability very close to one (13). In this study both LASSO and MCP models were fitted using the ncvreg package in the R 3.3.2 software.
2.3. Random Forest Classification with Variable Selection
Random Forest (RF) is one of the well-known machine learning methods consisting of several decision trees. Random Forest, which was proposed in 2000 by Brieman, for classification tasks, assigns a new observation to a class with the major votes (5). In this study, the researchers used the Gini index criteria to determine the splitting in each tree and 500 trees were considered to construct the forest. Moreover, in order to select the variables from random forest, the researchers eliminated the least important biomarkers successively based on out of bag error as minimization criterion using the varSelRF package in the R 3.3.2 software.
3. Results
The mean age of the participants in the control group was 34.5 and that of the patients in the case group was 33, which was not significantly different (P = 0.32). Also, in terms of demographic variables, such as gender, the 2 groups were the same. In order to assess the effects of the potential biomarkers in IgA nephropathy, first of all, the researchers performed Mann Whitney test as a univariate analysis and showed that the case and control groups had a significant (P < 0.05) difference in 144 out of 493 proteins. Because the sample size was even smaller than significant biomarkers, logistic regression analysis could not be applicable as a multivariate analysis. Therefore, the nature of the problem caused the researchers to use penalized regression methods, which are considered as a modern variable selection technique.
At the initial stage, amounts of shrinkage in both LASSO logistic and MCP logistic regression were estimated using 5-fold cross validation method as 0.023 and 0.025 for LASSO and MCP, respectively.
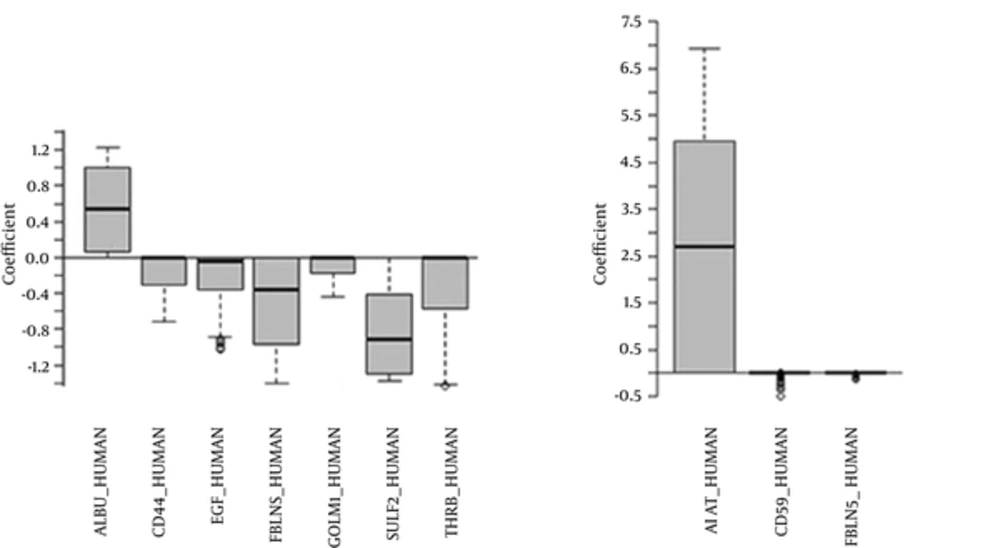
The LASSO logistic regression revealed that 7 out of 493 potential biomarkers had a significant contribution in the IgAN diseases. The most effective biomarker was SULF2 (OR = 0.28), followed by ALBU (OR = 2.66) and CD44 (OR = 0.97). Also, MCP logistic regression showed only 3 biomarkers as effective factors in IgAN diseases. The identified biomarkers in MCP model were A1AT (OR = 73.7), CD59 (OR = 0.95), and FBLN5 (OR = 0.95) (Table 1). The non-zero coefficients of both models were extracted using 500 times bootstrap method and shown in Figure 1.
| LASSO Logistic Regression | MCP Logistic Regression | Random forest | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Biomarker | Beta | SE | OR | Biomarker | Beta | SE | OR | Biomarker | MDA |
| SULF2 | -1.27 | 0.62 | 0.28 | A1AT | 4.30 | 2.51 | 73.7 | GOLM1 | 0.017 |
| ALBU | 0.98 | 0.58 | 2.66 | CD59 | -0.05 | 0.28 | 0.95 | IBP7 | 0.012 |
| THRB | -0.73 | 0.43 | 0.48 | FBLN5 | -0.05 | 0.74 | 0.95 | ALBU | 0.011 |
| FBLN5 | -0.39 | 0.57 | 0.68 | - | - | - | - | APOE | 0.011 |
| EGF | -0.38 | 0.26 | 0.68 | - | - | - | - | CD44 | 0.011 |
| GOLM1 | -0.14 | 0.23 | 0.87 | - | - | - | - | - | - |
| CD44 | -0.03 | 0.39 | 0.97 | - | - | - | - | - | - |
Selected Biomarkers in the Three Proposed Models
In random forest method, 5 biomarkers with higher mean decrease accuracy (MDA) were selected as the only effective factors on IgAN. GOLM1 and IBP7 were the most important biomarkers in RF with 0.017 and 0.012 mean decrease accuracy, respectively (Table 1).
In terms of prediction accuracy, all of the 3 proposed models were able to truly differentiate all the IgAN patients from the control groups so the area under the ROC curve, sensitivity and specificity were 100%. Moreover, in the LASSO model, the range of probability of IgAN was 0.94 to 0.99 for the case group and 0.01 to 0.04 for the control group and optimum cutoff point was obtained using the ROC analysis as 0.49 for probability of IgAN. Besides, the optimum cutoff point was obtained as 0.41 in the MCP model and the range of probability of IgAN was 0.76 to 0.90 and 0.00 to 0.06 for the case and control groups, respectively.
4. Discussion
To identify the most effective biomarkers on IgA nephropathy, the researchers applied LASSO, MCP, and random forest in their high dimensional proteomic data set. Comparison of the total number of selected biomarkers (11 biomarkers) with the protein profile (493) indicated that as expected, most of the biomarkers had no role in diagnosis. In terms of prediction accuracy, the proposed models were the same because they truly differentiated all the IgAN patients from the control groups.
Selected biomarkers among the 3 models were somewhat different, which may be due to the small sample size. Compared to random forest (machine learning techniques generally), LASSO and MCP represent more stable results, where the number of biomarkers is much larger than the sample size (14, 15). Moreover, penalized methods are more interpretable than machine learning techniques using concepts like odds ratio for biomarkers and probability of disease for each patient. One the other hand, random forest, as one of the most powerful machine learning techniques, is not restricted to linear associations and could detect any relationship between biomarkers and disease (5).
Since the sensitivity and specificity of the 3 models were 100%, all the eleven significant proteins are important in disease development. Nevertheless, the researchers suggest that the most important urinary proteins that have the highest diagnostic value are more suitable for further validation, whether the protein ID was significant in at least 2 models. Accordingly, the suggested panel is composed of: FBLN5, GOLM1, and CD44. The researchers excluded albumin, because it is non-specific and is excreted in the urine in all types of kidney diseases with proteinuria; however, the fragments of the excreted albumin and the amount of excretion might be disease-specific. Therefore, the significance of albumin, as one of the most abundant proteins in the serum and urine in this condition seems to be logical, and needs to be considered in future experiments for exploring the excretion of the specific fragments of albumin in IgAN.
All of these 3 suggested diagnostic biomarkers are down-regulated in IgAN patients in the present dataset with a fold change of 11.3, 2.4, and 10.6 for FBLN5, GOLM1, and CD44, respectively.
Furthermore, FBLN5, as a multifunctional glycoprotein, belongs to the fibulin family that has been reported to have a relationship with elastic system fibers, and contributes in assembly and organization of the extracellular matrix and regulation of microfibril formation (16-18). Decreased excretion of FBLN5 that is also known as fibulin-5 in IgA nephropathy patients compared with healthy individuals indicates the impairment of elastic system fibers and extracellular matrix (ECM)-cell interaction in this disease. A hypothesis on the relationship between decreased urinary excretion of FBLN5 and IgA nephropathy is accumulation of microfibrils and aberrant remodeling of the ECM in expansion of mesangial matrix that are mediated by FBLN5 and cause a decrement in urinary FBLN5 compared with normal conditions.
Furthermore, GOLM1 is a cis-Golgi membrane protein with unknown function (19). This protein is a known non-invasive biomarker for prostate cancer (20), which is predominantly expressed by the cells of the epithelial lineage, especially in the liver and kidney (21). Defects in GOLM1 gene leads to the development of renal disease, most notably focal segmental glomerulosclerosis and hyaline thrombi (22). This is the first time that GOLM1 is suggested as a potential biomarker of IgA nephropathy. Further experiments are essential for validation of urinary changes of GOLM1 in IgAN patients in a larger cohort.
The third suggested biomarker for IgAN, CD44, is also involved in cell-cell and cell-matrix interactions (23). The CD44 is a marker of activated Parietal Epithelial Cells (PECs) (24), whose expression is markedly enhanced in inflammatory renal diseases (25). Over-expression of CD44 in the renal tissue of IgA nephropathy patients was previously reported by Kim et al. and Florquin et al. (26, 27). They reported a positive correlation between CD44 expression and degree of proteinuria as well as degree of renal damage (26). There is evidence on decreased urinary excretion of CD44 in advanced stage of IgA nephropathy compared with the primary stage (28). However, decreased urinary excretion of this protein was significant in the current study and helped to discriminate the case from the control group. The different pattern of changes of this biomarker in different studies might be due to different samples: tissue versus urine (10).
4.1. Conclusion
Because all the 3 models were able to truly differentiate all the IgAN patients from the control groups, the researchers suggest that the proposed model could be used for modeling high dimensional and low sample size datasets.