1. Background
Achondroplasia (OMIM# 100800) is the most common skeletal dysplasia associated with disproportionate short stature. The prevalence of achondroplasia is about 1:10,000 to 1:30,000 live births (1). Meta-analysis showed a higher prevalence of achondroplasia in North Africa and the Middle East, about 34.31 per 100,000 people, compared to other regions such as Europe and America (2). Achondroplasia is inherited in an autosomal dominant pattern; about 80% of cases are caused by new mutations. Achondroplasia is caused by gain-of-function mutations in the FGFR3 gene located at the 4p16.3 locus (3, 4). The mature FGFR3 protein is a membrane receptor tyrosine kinase with an extracellular binding domain, which consists of three subdomains of immunoglobulin in the extracellular part, a membrane domain, and intracellular tyrosine kinase domains (5, 6). In the normal state, FGFR3 is bound to different fibroblast growth factors (FGFs) and is activated. This binding appears to result in dimerization of the receptor, activation of tyrosine kinase, and phosphorylation of the tyrosine residues. These changes lead to the activation of several downstream signaling pathways, including STAT and MAPK. In general, these secondary pathways slow down the proliferation and differentiation of chondrocytes. A mutation in FGFR3 stimulates and stabilizes the dimerization of the receptor and the permanent activity of FGFR3 even in the absence of FGF ligand binding. This permanent activity can cause continuous activation of the aforementioned signaling pathways such as MAPK, STAT1, and PLCγ, which ultimately disrupts the proliferation and differentiation of chondrocytes and leads to inhibited bone growth. The substitution mutation c.1138G>A in the FGFR3 gene is responsible for 97 - 98% of disease cases. This mutation leads to substituting glycine with arginine at position 380 in the transmembrane domain of FGFR3 (7, 8). Achondroplasia is characterized by distinct craniofacial features like macrocephaly, frontal bossing, and middle face hypoplasia, disproportionate short stature with rhizomelic shortness of limbs, brachydactyly, kyphosis, limited elbow movement, genu varum, and trident hands appearance. The average expected height of adults in men is 131 cm and 125 cm in women (9). The diagnosis of achondroplasia is based on clinical manifestations and radiographic findings. There are several skeletal dysplasias caused by mutations in the FGFR3 gene, such as thanatophoric dysplasia and hypochondroplasia. These diseases make the final diagnosis of achondroplasia challenging due to the overlap of clinical symptoms and radiological results. Therefore, the definitive diagnosis can be verified only by performing a genetic analysis of the FGFR3 gene and identifying the disease mutation (10). Definitive diagnosis of achondroplasia by genetic testing is very important because it not only can lead to a timely diagnosis, which can be very effective in improving the quality of life of patients, but also can be very helpful in prenatal diagnosis and genetic counseling (11). Therefore, finding a suitable genetic diagnostic method is essential. ARMS-PCR is a fast and inexpensive technique that can detect point mutations with high accuracy. Considering that the point mutation c.1138G>A is observed in about 97 - 98% of achondroplasia patients, this technique can identify a significant portion of patients (12).
2. Objectives
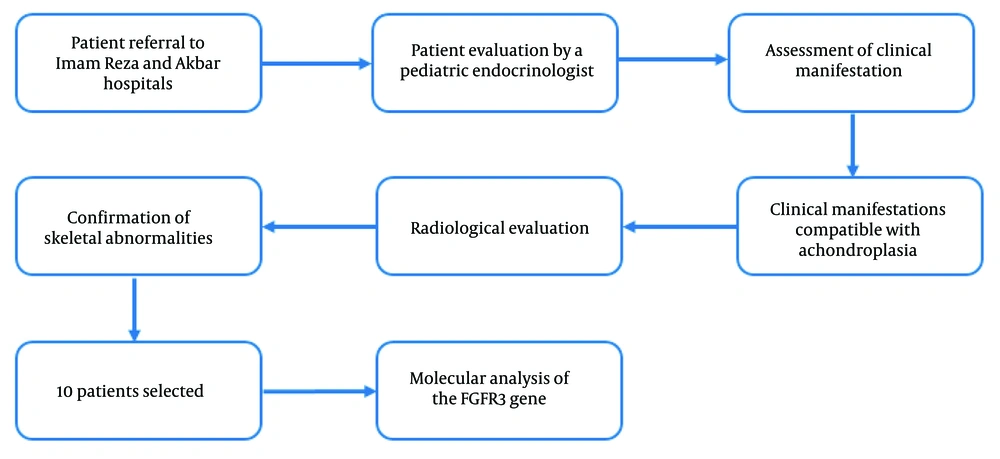
In this study, we examined 10 achondroplasia patients from Khorasan Razavi province, who were referred to Akbar and Imam Reza hospitals. Initially, all patients were analyzed for the presence of the common mutation using ARMS-PCR. Then, the exonic and adjacent intronic regions of FGFR3 in patients without the common mutation were examined by Sanger sequencing. The purpose of the study is to genetically evaluate achondroplasia patients to identify the disease mutation and also introduce ARMS-PCR as a convenient and fast technique for the final and definitive diagnosis of achondroplasia.
3. Methods
3.1. Subjects
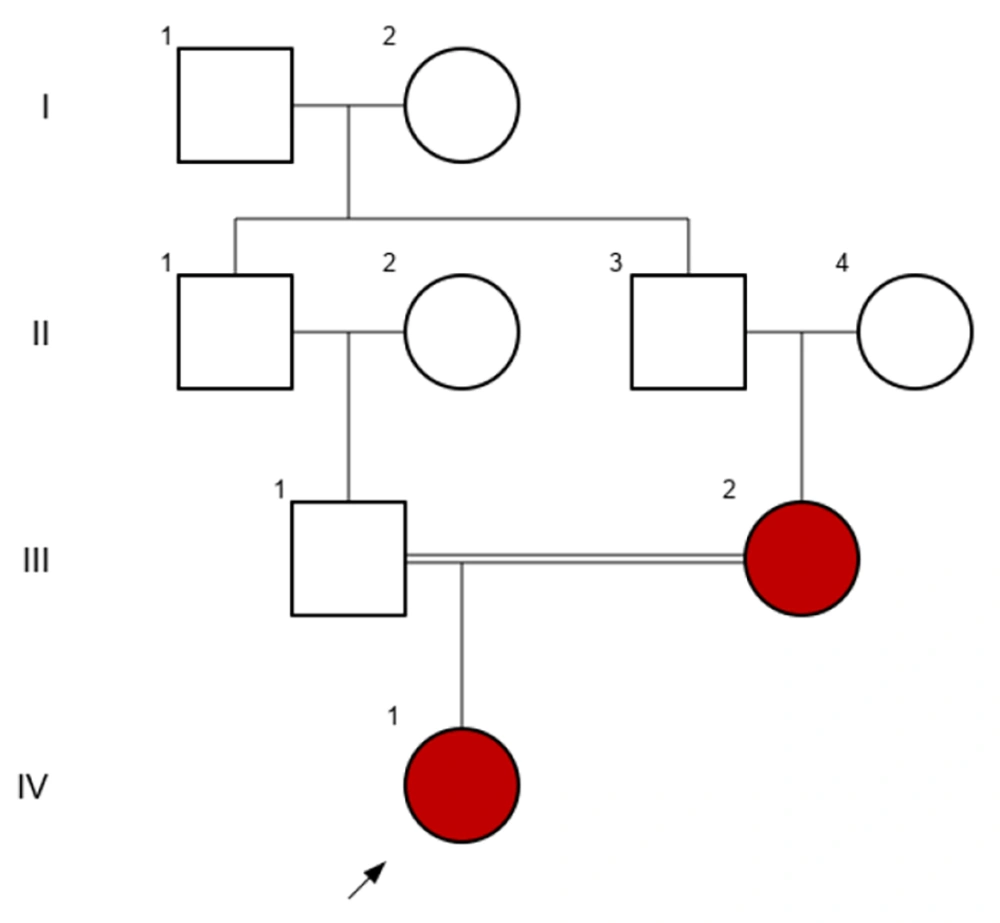
This retrospective study investigated 10 patients diagnosed with achondroplasia who were referred to Imam Reza and Akbar hospitals in Mashhad between 2018 and 2023. Patients were chosen based on their availability and hospital referral, as well as their diagnosis during the course of the study. All patients were evaluated by a pediatric endocrinologist to ensure appropriate patient selection for molecular analysis. Clinical symptoms and radiographic imaging, with attention to the pediatric endocrinologist's opinion, were the inclusion criteria of the study, and all participants met the following characteristics. Clinical symptoms included disproportionate short stature and skeletal features such as macrocephaly, prominent forehead, trident hands, and brachydactyly. Additionally, radiographic findings were utilized for diagnosis, which included a relatively large cranial vault with a small skull base, posterior vertebral scalloping, anterior flaring of the ribs, small squared iliac wings, and shortened tubular bones with characteristic bowing (Figure 1). The patients' ages ranged from 3 to 18 years. After receiving explanations, patients and their parents knowledgeably signed the consent form to participate in this research, and their clinical information was collected (Table 1). Subsequently, the INVITAE online program was utilized to record the patients' family histories and create pedigrees for their families. In Figure 2, the pedigree of a patient with a family history of achondroplasia is shown.
| Patients | Gender | Short Limb and Stature | Father's Age | Family History | Craniofacial Features | Brachydactyly | Genu Varum | Motor Development | Complications | Radiographic Images | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Macrocephaly and Frontal Bossing | Midface Retrusion | ||||||||||
| P1 | Female | + | 30 | - | + | + | + | - | Delay | + | |
| P2 | Male | + | 38 | - | + | + | + | - | Delay | + | |
| P3 | Female | + | 25 | + | + | - | + | - | Delay | Kyphosis | + |
| P4 | Female | + | 31 | - | + | + | + | + | Delay | + | |
| P5 | Female | + | 35 | - | + | - | + | - | Normal | Kyphosis | + |
| P6 | Female | + | 35 | - | + | + | + | - | Normal | + | |
| P7 | Female | + | 33 | - | + | + | + | - | Delay | + | |
| P8 | Male | + | 28 | - | + | + | + | + | Delay | Recurrent otitis media | + |
| P9 | Female | + | 37 | - | + | + | + | - | Delay | + | |
| P10 | Male | + | 41 | - | - | + | + | - | Delay | + | |
3.2. DNA Extraction
Peripheral blood samples of the participants in this study were collected in tubes with EDTA anticoagulant. Following the standard procedures in molecular genetics labs, DNA was extracted using the salting-out method.
3.3. Primer Design
Genotyping of the c.1138G>A mutation was conducted using the ARMS-PCR method. This technique employs four primers, using amplicons that correspond to both allelic forms and a larger fragment of DNA including the substitution mutation. A mismatch is inserted at the 3' end of each of the two allele-specific primers to enhance the specificity of the reaction. The GeneBank accession number for FGFR3 was used to guide the design of primers. We utilized the "BLAST" program to determine the primers' specificity. Additionally, important parameters of primers, including PCR product length, GC percentage, annealing temperature, and the possibility of secondary structure formation, were checked using the online database IDT and Oligo Analyzer tool. The primers used in this study were: Forward outer c.1138G>A: 5'-GTTACTGACTGCGAGACCCT-3'; reverse outer c.1138G>A: 5'-GTTTCGTGCCCCAAAGTACC-3'; forward inner c.1138G>A: 5'-CAGGCATCCTCAGCTCCA-3'; and reverse inner c.1138G>A: 5'-AACAGGAAGAAGCCCACACC-3'. Pishgam Biotechnology Company was then notified for primer synthesis orders.
3.4. PCR Method
A total of 20 µL of PCR mixture comprising 20 - 100 ng of DNA, 10 µL of 2X Master mix (Pars Tous, Ref. No. 101081, Iran), 4 µL of 5X Q-solution, 1 µmol of 5 pM of each primer, and 1 - 2 µL of distilled water were used in the process. The thermocycler machine (SensoQuest GmbH, Germany) was used for the PCR. The protocol included a 5-minute initial denaturation step at 95°C, 32 cycles of 30 seconds at 95°C, 30 seconds at 59°C for the A allele and 60°C for the G allele, 30 seconds at 72°C, and a 5-minute final extension step at 72°C. Five microliters of the PCR product were electrophoresed at 100 V on 2% standard agarose gels for 50 minutes. Ethidium bromide was used to visualize the fragments using an ultraviolet transilluminator.
3.5. Validation of c.1138G>A Detection by Sanger Sequencing
As previously mentioned, PCR was performed with c.1138G>A forward outer and c.1138G>A reverse outer primers. Codon Company sequenced PCR products using the c.1138G>A reverse outer primer. Sanger sequencing was performed on five patients. Of these, three patients revealed a 318 bp amplicon corresponding to the mutant allele, while two patients lacked a 318 bp amplicon. Using a 2X master mix, the PCR reactions were first primed (Pars Tous, Ref. No. 101081, Iran) with appropriate primers (5 picomolar) in a thermocycler machine (SensoQuest GmbH, Germany) for 35 cycles at a specific temperature. After PCR product specificity was confirmed and electrophoresis on a 2 percent agarose gel, the products were sent to Codon Company for Sanger sequencing. UGENE software was used to visualize all sequencing data and compare them to reference sequences found in the NCBI database. The detected variants were analyzed in disease-related databases like ClinVar, HGMD, OMIM, and VarSome to ascertain their clinical significance.
4. Results
4.1. Identification of Common Mutation c.1138G>A Using ARMS-PCR
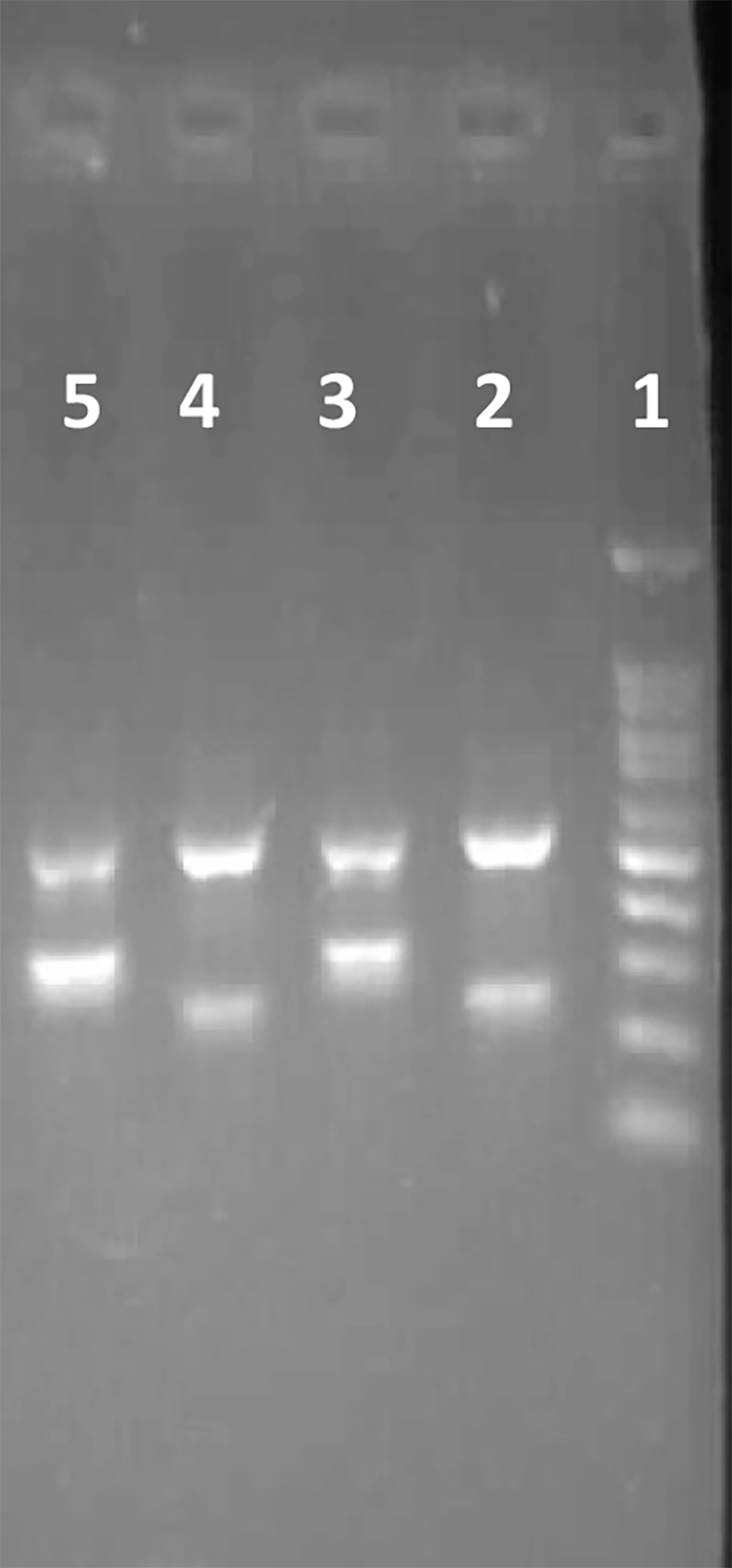
In an analysis of 10 patients with achondroplasia, we conducted genetic testing to identify potential mutations in the FGFR3 gene, which has been strongly associated with the condition. We employed an assay that is straightforward, rapid, and reliable for the accurate and timely identification of the mutant c.1138G>A allele. In the ARMS-PCR assay, two primers specific for the allele-specific amplification of the wild-type allele and the mutant allele were combined in two distinct tubes with two primers complementary to distinct sequences of FGFR3. Numerous elements that can impact the specificity and efficiency of PCRs, such as primer concentration and PCR cycling conditions, were optimized. Annealing temperatures between 57 and 63°C were tested. During the PCR reaction, a 517 bp region of FGFR3 was amplified using c.1138G>A forward outer and c.1138G>A reverse outer primers. This region functioned as a template for the ensuing allele-specific amplification and an internal control for the PCR amplification's quality. The 236 bp PCR product specific for the wild-type allele and the 318 bp PCR product specific for the mutant allele were obtained from the allele-specific amplification (c.1138G>A reverse inner/c.1138G>A forward outer and c.1138G>A forward inner/c.1138G>A reverse outer). The presence of the 318 bp amplicon indicates a positive result; therefore, the presence of the c.1138G>A mutation in the patient. The 236 bp amplicon indicates amplification of the wild-type allele. Given that achondroplasia is an autosomal dominant condition and its homozygous status is fatal, it is expected that patients carrying the c.1138G>A mutation will have both the 318 bp and 236 bp amplicons alongside the 517 bp amplicon. The absence of the 318 bp amplicon indicates the lack of the c.1138G>A mutation in the patient, necessitating further investigation of other exons and adjacent intronic regions to identify the disease-causing mutation. A representative ARMS-PCR assay analysis is shown in Figure 3. According to the results of this study, the common mutation c.1138G>A was identified in eight of the examined patients. Figure 4 shows the validation of the presence and absence detection of the common mutation c.1138G>A with ARMS-PCR by Sanger sequencing. Based on the consistency of ARMS-PCR results with Sanger sequencing, it can be concluded that the specificity of ARMS-PCR in identifying the c.1138G>A mutation is high and reliable.
Evaluation of the patients with agarose gel electrophoresis of the ARM-PCR products. 1, 100 bp DNA ladder; 2, 3, patient 1; and 4 and 5, patient 2 both carry the c.1138G>A mutation; 2 and 4, wild-type allele (236 bp) and internal control (517 bp); 3 and 5, mutant allele (318 bp) and internal control (517 bp).
4.2. Identification of c.1620C>G in Two Patients
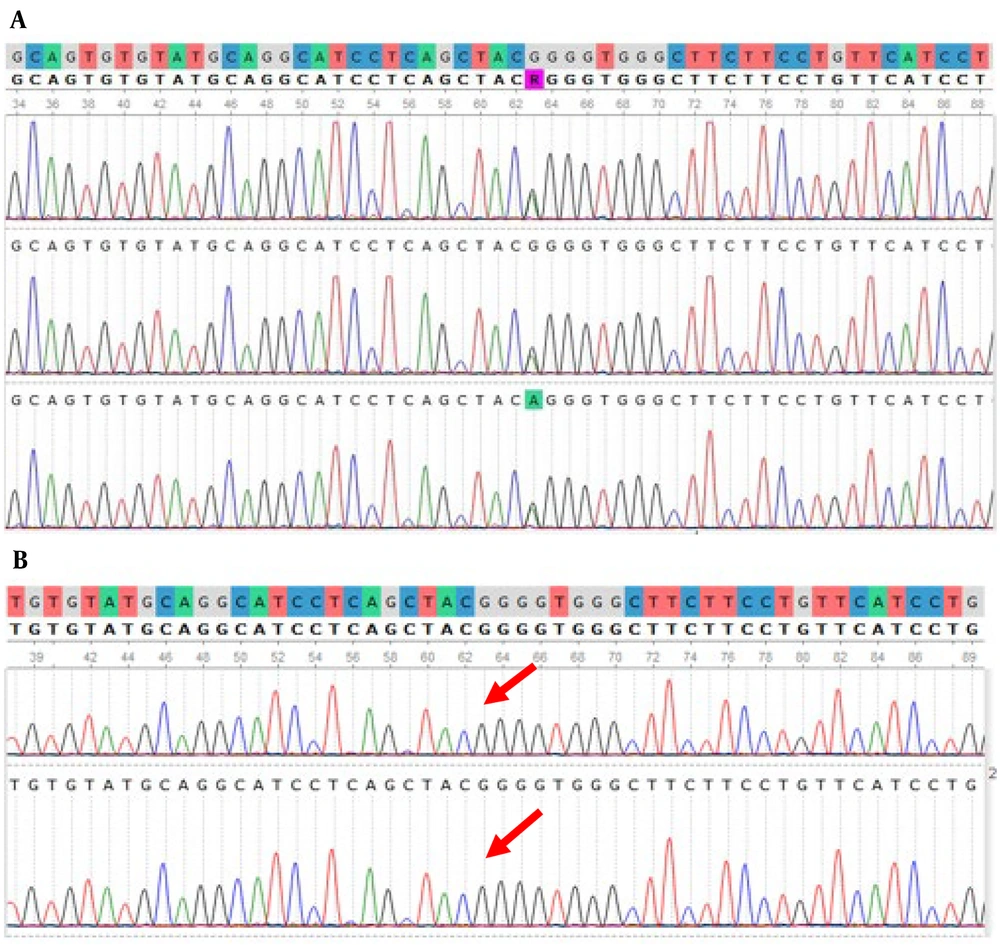
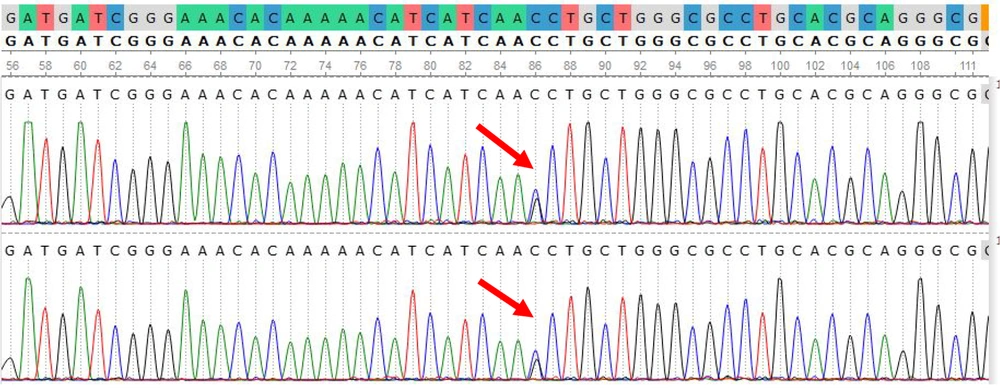
Our study revealed that out of the 10 patients, two did not indicate a 318 bp amplicon, prompting further examination of the other exons of FGFR3 for these individuals. They exhibited a specific mutation, c.1620C>G, in the FGFR3 gene. The c.1620C>G mutation is one of the well-documented alterations in this gene, substituting an asparagine residue with lysine at position 540 of FGFR3. This mutation is more related to hypochondroplasia, which is very similar to achondroplasia but has milder phenotypes compared to it. The c.1620C>G mutation is one of the most common mutations of hypochondroplasia and is found in 50 - 70% of hypochondroplasia patients. Figure 5 represents the identified mutation in the FGFR3 gene, as viewed by UGENE. Table 2 shows the result of the molecular analysis of the FGFR3 gene in 10 patients.
| Patients | Detecting c.1138G>A Mutation by ARMS-PCR | Detecting Mutation in Other Exons | Zygosity | |
|---|---|---|---|---|
| Exon | Mutation | |||
| P1 | + | - | - | Heterozygous G/A |
| P2 | + | - | - | Heterozygous G/A |
| P3 | - | 12 | c.1620C>G | Heterozygous C/G |
| P4 | + | - | - | Heterozygous G/A |
| P5 | + | - | - | Heterozygous G/A |
| P6 | + | - | - | Heterozygous G/A |
| P7 | + | - | - | Heterozygous G/A |
| P8 | + | - | - | Heterozygous G/A |
| P9 | + | - | - | Heterozygous G/A |
| P10 | - | 12 | c.1620C>G | Heterozygous C/G |
5. Discussion
The final diagnosis of achondroplasia is only made through genetic evaluation and identification of disease mutations. Since accurate and timely diagnosis is beneficial in managing disease complications and improving patients' quality of life, as well as in genetic counseling, genetic evaluation of achondroplasia and finding a good technique for diagnosis is crucial (13, 14). Several studies have been conducted with the aim of genetic investigation of achondroplasia in different countries. For example, Heuertz et al. in France conducted genetic evaluations on a series of 75 achondroplasia and hypochondroplasia patients. They observed mutations that lead to an increase in cysteine residues, resulting in a more severe phenotype. They recommended genetic examination for the final diagnosis (15, 16). Bucerzan et al. in Romania studied 27 achondroplasia patients and observed the common mutation c.1138G>A in only 16 cases, although all patients had been diagnosed with achondroplasia based on clinical and radiographic results (6). In China, a study by Zhang et al. was conducted on 17 children with achondroplasia. The notable point of this study was the identification of two unreported variants, c.1252C>T and c.445+2-445+5delTAGG in FGFR3; while the clinical phenotype of these two patients was not different from the others with common mutations (17). To our knowledge, no study has been done in Iran so far; therefore, in this study, we conducted the genetic evaluation of 10 achondroplasia patients. In 8 patients, the common mutation c.1138G>A was detected, and in 2 patients, the mutation c.1620C>G, which is the most common mutation associated with hypochondroplasia, was detected (18). While all patients were eligible for inclusion criteria, containing clinical symptoms and radiographic imaging, all of them were evaluated comprehensively by a pediatric endocrinologist, and there was no remarkable difference in their clinical features. Our results indicate the overlap of achondroplasia manifestation with hypochondroplasia, like other studies, and confirm the considerable role of genetic examination in the final diagnosis. Additionally, finding an appropriate technique for identifying achondroplasia patients is necessary. Different methods have been investigated to find a suitable diagnostic (19). For example, Patil et al. and Pehlivan et al. both used PCR-RFLP to identify patients with the mutation c.1138G>A and showed that PCR-RFLP identified the patients appropriately (20, 21). He et al. examined PCR-RFLP and HRM analysis simultaneously to find patients with common mutations at position 1138. According to the similarity of the results, they presented that HRM analysis is a more suitable option for diagnosing (22). Su et al. investigated the DHPLC technique intended for rapid identification of achondroplasia patients and introduced it as a technique with good sensitivity and accuracy for clinical application (23, 24). Although several techniques have been proposed for diagnosis, for using a technique in clinical cases and on a large scale, it is required that the technique be easy and affordable. DHPLC is a good technique with acceptable results for common mutation identification responsible for achondroplasia, but the weakness of DHPLC in DNA analysis is an obstacle to its application in clinical cases (25). HRM analysis and DHPLC require specialized facilities. Also, the interpretation of melting curves in HRM is challenging, particularly in heterozygous patients like those with achondroplasia, due to the presence of several peaks; besides, sequencing is required for result confirmation (26, 27). The possibility of incomplete digestion of PCR products that can lead to false positive results is a weakness of PCR-RFLP (28). Hence, there is still a need to find a suitable genetic diagnostic method for achondroplasia. In this study, we used ARMS-PCR to identify patients with the c.1138G>A mutation (29). The validation of ARMS-PCR results with Sanger sequencing, considering that Sanger sequencing is the gold standard for validating specific genetic variants, indicates that ARMS-PCR can identify achondroplasia patients with the common mutation with 100% accuracy at a low cost. In addition, performing ARMS-PCR does not require expensive facilities or reagents, which makes it applicable in various laboratories (30). Furthermore, ARMS-PCR is an easy and fast genetic test; its results are prepared in a few hours, making it suitable for clinical practice. The outer primers used in this method act as an internal control and confirm the PCR reaction, which is an important merit over other PCR-based methods (31, 32). Therefore, while other assays, including PCR-RFLP, HRM analysis, and DHPLC, have reported high accuracy (100%) and sensitivity for identifying the common mutation, it can be argued that ARMS-PCR outperforms these techniques in larger scale and clinical applications due to its characteristics like rapidity, cost-effectiveness, simplicity, and no requirements for specialized equipment and expertise.
Despite promising results, our study has some limitations. One of the important limitations is that our study patient group is not large enough for the confirmation of ARMS-PCR usage in clinical cases. Due to achondroplasia being a rare condition and limitations in time and resources, we could investigate only ten patients, and further investigation was impossible. Another limitation was the restriction in sample collection from other centers on a larger scale, which could affect the generalizability of our results and the chance of finding other disease-causing mutations in different populations. For understanding the mutation distribution of the FGFR3 gene for achondroplasia in different populations, as well as the clinical utilization of ARMS-PCR, more studies within larger-scale cohorts are suggested.
5.1. Conclusions
In this study, we investigated the genetic aspect of achondroplasia and showed that due to the overlap of clinical manifestations and radiographic findings with other skeletal dysplasias, especially hypochondroplasia, the definitive diagnosis of achondroplasia, which is crucial, can be done only using molecular examination and identification of the mutation. Additionally, in this study, we used ARMS-PCR for the rapid identification of achondroplasia patients with the common mutation, c.1138G>A, which is observed in 97 - 98% of patients. We indicated that ARMS-PCR can identify most achondroplasia patients accurately and easily with reproducible results.