1. Background
Alpha-Thalassemia (-thal), one of the most common genetic disorders in the world, is characterized by deficient (+) or absent () synthesis of the alpha chain of the hemoglobin (Hb) molecule (1). Most -thal determinants are deletions involving one or both of the duplicated alpha-globin genes. However, non-deletion -thal mutations have been reported, which include termination codon mutations such as Hb Constant Spring (2), splicing defect caused by a 5-basepair (bp) deletion of the first intervening sequence (3), Hb Quong Sze, extremely unstable a-globin structural variant (4, 5), single nucleotide substitution of the polyadenylation site (6), two nucleotide deletions at position -1 and -2 of the 5' untranslated region preceding the AUG codon (7), initiation codon mutation (AUG-ACG) (8) and nonsense mutation (a 116 GAG-UAG) (8). Apart from the -1, -2 deletion that occurs in a single a-globin gene chromosome, each of these mutations affects the 2-globin gene.
2. Objectives
Here, we report on a novel mutation in two unrelated families with moderate microcytosis aneamia. Sequencing of the 2 and 1 globin genes revealed a novel heterozygous point mutation at the 5'UTR region of the 2-globin gene.
3. Materials and Methods
3.1. Patients
Among patients referred to the Genetic Antenatal Clinic at the Ghaem Hospital of Mashhad, two families showed an inherited novel mutation. Prior to any laboratory work, according to the ethics community, a written consent was obtained from all patients. In the first family, the proband (II-1) and her parents (I-1 and I-2) were studied (Table 1). The proband (II-1) and her mother (I-2) carried the mutation while the father (I-1) had a normal genotype. In the case of the second family (Table 2) the proband (II-1) and his mother (I-2) carried the mutation while his sister (II-2) had a normal genotype. The red blood cell (RBC) indices, and Hb A2 and Hb F percentages were determined and alkaline electrophoresis on cellulose acetate and citrate agar electrophoresis were carried out according to standard procedures (9). The red cell lysates were also analyzed by isoelectric focusing (IEF) and exchanges high performance liquid chromatography (HPLC) (10).
| PARAMETERS | II-1 | I-1 | I-2 |
|---|---|---|---|
| SEX-AGE, Y | F-16 | M-42 | F-36 |
| GENOTYPE | ΑΑ/ΑTΑ | ΑΑ/ΑΑ | ΑΑ/ΑTΑ |
| RBC, 1012/L | 5.17 | 7.03 | 5.32 |
| HB, G/DL | 13.2 | 17.5 | 13.2 |
| MCV, FL | 78.3 | 80.0 | 79.5 |
| MCH, PG | 25.5 | 27.0 | 24.8 |
| HBA2, % | 2.5 | 2.7 | 2.0 |
| HBF, % | 1.9 | 0.6 | 1.6 |
| FERRITIN (ECL) | 31.2 | 37 | 15.4 |
| Β-GENOTYPE (SEQUENCING) | NORMAL | NORMAL | NORMAL |
| Parameters | II-1 | II-2 | I-2 |
|---|---|---|---|
| Sex-age, y | M-24 | F-32 | F-68 |
| Genotype | αα/αTα | αα/αα | αα/αTα |
| RBC, 1012/L | 6.06 | 4.71 | 4.67 |
| Hb, g/dL | 14.9 | 13.4 | 12.8 |
| MCV, fL | 73.8 | 90.0 | 71.2 |
| MCH, pg | 24.6 | 28.5 | 23.2 |
| HbA2, % | 3.0 | 2.5 | 2.4 |
| HbF, % | 1.2 | 1.0 | 1.4 |
| Ferritin, ECL | 26 | 54 | 13 |
| β-Genotype (sequencing) | Normal | Normal | Normal |
3.2. Molecular Analysis
For molecular analysis genomic DNA was isolated from peripheral blood cells by a salting out procedure (11). The common alpha deletion mutations were ruled out using the published primers and conditions (12). The amplification of the entire and 1 globin genes was also carried out and their DNA was sequenced. No was mutation detected (the primers and PCR conditions are available upon request). The 2-globin gene was amplified in an Applied Biosystems 2720 thermal cycler using 10 pmol of the following primers: forward (5΄- GCTCCGCGCCAGCCAATGAG -3΄); reverse (5΄- GAGAGGTCCTTGGTCTGAGACAG -3΄) using Hot star Taq DNA polymerase (Qiagen). The 2-globin gene was sequenced in both forward and reverse directions using an ABI PRISMTM 310 Genetic Analyzer (PE Applied BioSystems, Foster City, CA, USA) and the ABI PRISM Big Dye Terminator v3.1 Cycle Sequencing Kit (PE BioSystems).
3.3. Gene Expression Analyses by Real-Time Polymerase Chain Reaction (PCR)
Cytoplasmic RNA was isolated by the use of QIAamp RNA Blood Mini Kit (Qiagen) according to manufacturer’s protocol. RNA was reverse transcribed by the use of oligo (dT) and superscript reverse transcriptase (Gibco-BRL). Real-time polymerase chain reaction (RT-PCR) was performed on the StepOne™ (48-well) Real-Time PCR system (Applied Biosystems) in MicroAmp™ Fast Optical 48-Well reaction plate (applied biosystems) with a total volume of 15 µL, containing cDNA (1.5 µL), forward and reverse primers (300 nM), SYBR® Premix Ex TaqTM (7.5 µL) (Takara, Japan) and ROXTM Reference Dye II (0.3 µL) (Takara, Japan) and dH2O 5.1 µL with Optical Adhesive Film (Applied Biosystems) in triplicates. Thermal cycling conditions were as follows: stage one, 95°C for 60 seconds; stage two (repeated 40 times), melting at 95°C for 10 seconds; stage three annealing and polymerizing at 60°C for 30 seconds. The relative standard curve method was applied for cDNA quantification. The quantities of the target genes were normalized with the quantity of beta-2 microglobulin (β2M) as the endogenous control gene. To obtain the final relative quantification (RQ), the normalized quantity of the unknown sample was compared with the normalized quantity of the control sample. Primer sequences were: β2M forward 5΄- GTA TGC CTG CCG TGT GAA C-3΄, reverse 5΄-AAC CTC CAT GAT GCT GCT TAC-3΄; alpha-2 (HBA2) forward 5΄-ACG CTG GCG AGT ATG GT -3΄, reverse 5΄- GCT TAG GAG CTT GAA GTT GAC -3΄.
4. Results
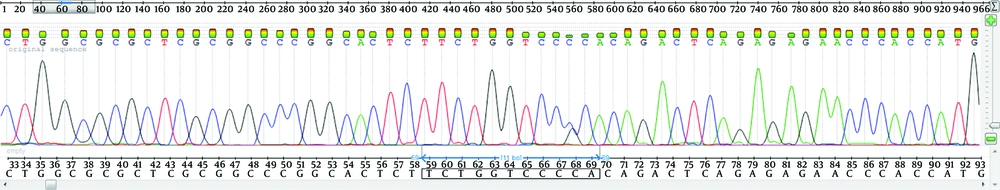
The hematological data of the probands and their families are presented in Tables 1 and 2. Hypochromia and microcytosis were found in the probands and in I-2 of both families. The father of the second family was deceased and the sister (II-2) did not carry the thalassemia mutation. No abnormal globin chain was detected by reversed phase HPLC. The common deletion mutations for 1 and 2 globin genes (12) and non-deletion 1 globin gene mutations and also anemia due to iron deficiency were ruled out. Molecular analysis of the beta-globin gene did not reveal any defect in any family member. In the probands and other affected members of the families, DNA sequencing of the 2 gene, showed a novel heterozygous C > G substitution, 23 nucleotides upstream of the ATG translation initiation codon. This was confirmed by reverse sequencing (Figure 1).
4.1. Alpha-2 Gene Expression
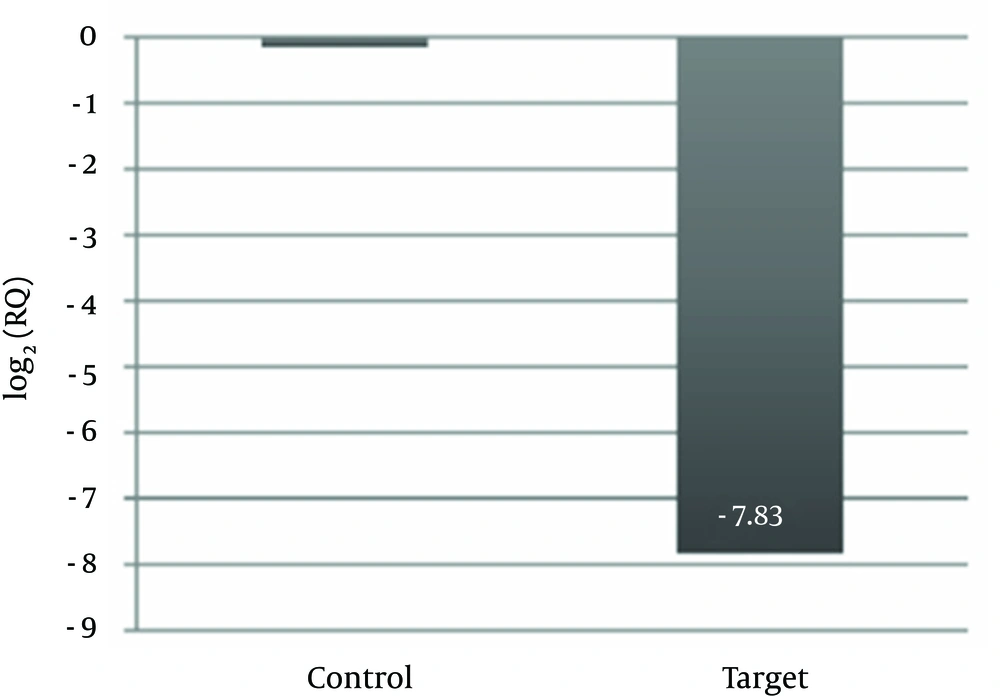
To evaluate whether this variant correlated with reduced mRNA levels, we measured α2-globin mRNA levels by real-time PCR. Real-time PCR results were analyzed automatically using the StepOne software v. 2.1 (Applied Biosystems). As shown in Figure 1, in this case, expression level of alpha-2 with respect to the control decreased dramatically. From our results a reduction of 277 folds is evident in alpha-2 gene expression (Figure 2).
5. Discussion
The mutation identified in our study was located on an AP-1 transcription factor binding site at position 222890 on chromosome 16. This mutation is within the 5'UTR region of the HbA2 gene. Although most mutations reported in the 5'UTR of the 2-globin gene are deletions (13-16) a T to G transversion in the regulatory binding motif of the -globin gene cluster can affect the regulation of gamma-globin gene expression (17). A high level of -globin expression depends on cis-acting regulatory sequences located upstream of the -globin gene. It has been shown that sequences containing the -globin positive regulatory element, activate globin expression in transgenic mice (18). The AP-1 transcription factor binding site in murine -globin contributes to long range -globin gene activation. Loyd et al. (19) demonstrated that a mutation in an AP-1/NFE2 transcription factor binding site motif could function over large distances and affect activation of - globin gene expression. They also demonstrated that the AP-1 binding sites are required for full enhancer activity and -globin gene transcription. Therefore, since a normal individual has four active α genes (αα/αα) and severity of α-thalassemia is related to the number of active α genes, fetuses with Hb Bart’s hydrop (−−/−−) died in utero. Patients with HbH disease have only one active α gene (−−/−α) and moderately severe but variable anemia. A person having two α genes (−α/−α or −−/αα) shows only a mild and sometimes asymptomatic form of α-thalassemia, whereas silent carriers (αα/−α) are generally normal. The disease is usually characterized by two main types; deletion HbH disease (−−/−α) and non-deletion HbH disease (−/−αTα).
The patients in this study had moderate microcytosis and hypochromia without hemolysis, jaundice and splenomegaly. Molecular analysis of these patients revealed a non-deletion type of mutation in the promoter region, which is highly consistent with the findings of previous studies (14-16).