1. Background
Flies are a large group of insects that are classified in the order Diptera and sub-order Cyclorrhapha or Muscomorpha (1). These insects have a high species diversity and about 150000 of their species have been identified and described in 158 families. Of these species, 285, 1500, 2500, and 5000 species belong to Fanidae, Calliphoridae, Sarcophagidae, and Muscidae families, respectively (2, 3). The medically important flies, in terms of close coexistence with humans and their dependence on human habitats, are known as domestic or synanthropic flies (4). They are also often called “Filth flies”; they include four species of Muscidae, Fanidae, Calliphoridae, Sarcophagidae families, and some similar flies (5). Flies are diurnal insects, which often live in and around the human environment. Although they are not major biologic vectors in the transmission of pathogens to humans, they can transmit several pathogens to their food and and environment because of their behavior, nutrition, and the power of flight and rapid displacement (6, 7). These insects contribute to the mechanical transmission of many pathogens to humans, especially in warm seasons. They are capable of transmitting more than 100 microorganisms of viral, bacterial, and parasitic pathogens, such as poliovirus, hepatitis A virus (HAV), hepatitis D virus (HDV), Chlamydia trachomatis, Coxiella burnetii, Cholera spp., Salmonella spp., Listeria spp., Streptococci spp., Staphylococci spp., Shigella spp., Entamoeba, Giardia, nematodes, and eggs of some tapeworms through the hair, mouthparts, legs, vomitus, and feces on foods. In addition, they can transmit some food-borne diseases to humans (8). On the other hand, the larvae of some flies can act as obligatory and facultative parasites of the vertebrate body tissues and cause myiasis in humans and animals (9). Previous studies have shown that the digestive tracts and the external body surfaces of the house flies (hairs, mouthparts, legs, vomitus, and feces) are contaminated with many human pathogens. Musca domestica is more important due to the fact that it coexists with humans (8). Animal husbandry in the rural areas of the Qom Province is a major occupation, and nearly 20000 households are involved in livestock breeding. On the other hand, the province has a plain and desert condition in terms of geologic pathology and has a warm and dry climate. The production of animal wastes in hot weather conditions provides good breeding places for flies and the early completion of their life cycle that can result in the generation of the flies.
2. Objectives
Therefore, this study was conducted to determine the bacteria contaminating Musca domestica (Diptera: Muscidae) collected from animal husbandries in the province of Qom, Iran, for the year 2019.
3. Methods
3.1. Study Sites
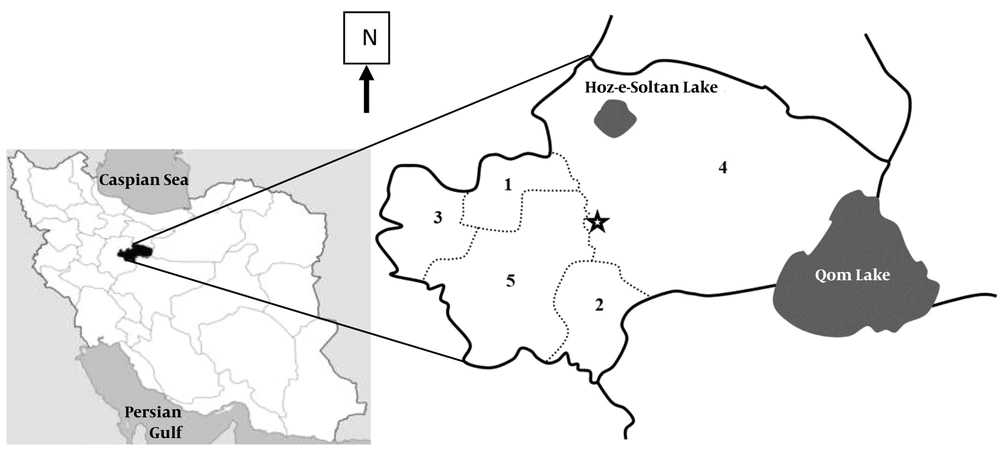
The Qom province is geographically located in the center of Iran (between 50 to 52 degrees east and 34 to 35 degrees north) (Figure 1). It has one urban area and five rural areas. Due to low altitude, small rainfall, inappropriate climate and saline lands, it is a part of the central desert of Iran. The Qom Province has a semi-desert climate and its average annual rainfall is less than 100 mm, which is mainly in autumn and winter. The rainfall in the warm seasons, especially in summer, is less than this amount (10). Therefore, it seems that the climatic and environmental factors are very suitable for the development of flies, especially house flies (11).
3.2. Flies Collection
To collect flies, one village and one urban texture were selected from each district; then, two animal husbandries were randomly chosen. They were sampled one time and a total of 160 flies were obtained. Capturing was done using a plastic water bottle fly trap (Figure 2) and insect net-hashing (Figure 3). The captured flies were then put into sterile glass containers. Finally, the samples were sent to the Department of Medical Entomology and Vector Control, School of Public Health, the University of Medical Sciences, Tehran, Iran.
3.3. Ethical Considerations
This study received financial support from the Deputy of Research, Qom University of Medical Sciences, Iran. Ethical clearance was earned from the Qom University of Medical Sciences, Iran (project no.: 95793, code of ethics: MUQ.REC. 1395.156).
3.4. Bacterial Cultures of Surfaces and Gut Contents of the House Flies
From each region, 20 house flies were individually evaluated. First, for the isolation of the bacteria from the external body of the flies, each one was poured in two tryptic soy broths (Merck, Germany). The media were mixed gently for five minutes, which resulted in the bound bacteria on the external surface of the insect to be released into the medium. Next, the sampled flies were transferred to new tubes to be disinfected by 70% alcohol for two minutes, followed by being washed by sterile normal saline. Then, they were dissected under the stereomicroscope by an entomologist and their digestive tracts were completely inoculated in two tryptic soy broths. Media were incubated at 37°C for 24 h (each of the media was incubated separately in ∼5% CO2 for fastidious bacteria).
3.5. Isolation and Detection of the Bacteria
For isolating both Gram-negative and Gram-positive bacteria, subculture from tryptic soy broth was done separately on blood agar, chocolate, eosin methylene blue agar (EMB), hektoen enteric agar (HEA), xylose lysine deoxycholate (XLD), lysine iron agar (LIA), salmonella-shigella agar (SSA), and mannitol salt agar (Merck, Germany). The media were incubated at 37°C for 24 h in ∼5% CO2. First, suspected colonies were evaluated by Gram staining. Then, a collection of the biochemical test was carried out for Gram-negative bacteria, such as catalase, oxidase, ortho-nitrophenyl-β-galactoside (ONPG), sulfide indole motility (SIM), methyl red (MR)/voges-proskauer (VP), citrate, urease, nitrate, oxidation/fermentation (OF), and fermentation of carbohydrate tests e.g. glucose, lactose, mannitol, sucrose, sorbitol, etc. The final confirmation of the Shigella spp. and Salmonella spp. was also done using specific antisera (Difco, USA). In positive cases of Shigella spp. and Salmonella spp., an agglutination reaction was observed between the antigen and antibody (12-14). Other biochemical methods (e.g., esculin hydrolysis, DNase, coagulase, catalase, hemolysis, growth on 6.5% NaCl, and susceptibility to optochin) were also used for the detection of the Gram-positive bacteria (15, 16).
4. Results
In total, 23 species from external surfaces (legs, hairs, and mouthparts) and from the digestive tracts of house flies, M. domestica, were isolated (Tables 1 and 2).
| Bacterial Species | Animal Husbandry | Total | |||
|---|---|---|---|---|---|
| Salafchegan | Khalajestan | Markazi | Kahak | ||
| Staphylococcus epidermidis | 16 (80) | 10 (50) | 4 (20) | 9 (45) | 39 (48.8) |
| Streptococcus pneumonia | 17 (85) | 8 (40) | 18 (90) | 17 (85) | 60 (75) |
| Staphylococcus aureus | 0 (0) | 0 (0) | 16 (80) | 14 (70) | 30 (37.5) |
| Enterococcus spp. | 0 (0) | 0 (0) | 0 (0) | 18 (90) | 18 (22.5) |
| Klebsiella oxytoca | 0 (0) | 0 (0) | 11 (55) | 0 (0) | 11 (13.8) |
| Proteus mirabilis | 0 (0) | 11 (55) | 17 (85) | 0 (0) | 28 (35) |
| Proteus vulgaris | 0 (0) | 12 (60) | 18 (90) | 0 (0) | 30 (37.5) |
| Serratia marcescens | 14 (70) | 20 (100) | 6 (30) | 16 (80) | 56 (70) |
| Escherichia coli | 20 (100) | 17 (85) | 18 (90) | 19 (95) | 74 (92.5) |
| Klebsiella pneumoniae | 0 (0) | 0 (0) | 14 (70) | 0 (0) | 14 (17.5) |
| Pseudomonas aeruginosa | 20 (100) | 20 (100) | 20 (100) | 20 (100) | 80 (100) |
| Providencia spp. | 11 (55) | 7 (35) | 8 (40) | 0 (0) | 26 (32.5) |
| Enterobacter aerogenes | 14 (70) | 13 (65) | 6 (30) | 11 (55) | 44 (55) |
| Shigella sonnei | 0 (0) | 0 (0) | 12 (60) | 19 (95) | 31 (38.8) |
| Salmonellaserogroup D | 0 (0) | 0 (0) | 14 (70) | 20 (100) | 34 (42.5) |
| Salmonella paratyphi A | 0 (0) | 0 (0) | 6 (30) | 0 (0) | 6 (7.5) |
| Shigella dysenteriae | 0 (0) | 0 (0) | 13 (65) | 20 (100) | 33 (41.3) |
| Citrobacter freundii | 0 (0) | 0 (0) | 0 (0) | 20 (100) | 20 (25) |
aValues are expressed as No. (%).
| Bacterial Species | Animal Husbandry | Total | |||
|---|---|---|---|---|---|
| Salafchegan | Khalajestan | Markazi | Kahak | ||
| Staphylococcus epidermidis | 20 (100) | 18 (90) | 10 (50) | 11 (55) | 59 (73.8) |
| Streptococcus pneumoniae | 10 (50) | 16 (80) | 9 (45) | 13 (65) | 48 (60) |
| Staphylococcus aureus | 11 (55) | 0 (0) | 0 (0) | 16 (80) | 27 (33) |
| Enterococcus spp. | 0 (0) | 11 (55) | 0 (0) | 0 (0) | 11 (13.8) |
| Klebsiella oxytoca | 8 (40) | 0 (0) | 8 (40) | 0 (0) | 16 (20) |
| Proteus mirabilis | 7 (35) | 0 (0) | 0 (0) | 0 (0) | 7 (8.8) |
| Proteus vulgaris | 12 (60) | 0 (0) | 0 (0) | 0 (0) | 12 (15) |
| Serratia marcescens | 15 (75) | 14 (70) | 13 (65) | 10 (50) | 52 (65) |
| Escherichia coli | 20 (100) | 20 (100) | 9 (45) | 10 (50) | 59 (73.8) |
| Klebsiella pneumoniae | 13 (65) | 0 (0) | 12 (60) | 0 (0) | 25 (31.3) |
| Pseudomonas aeruginosa | 0 (0) | 12 (60) | 7 (35) | 14 (70) | 33 (41.3) |
aValues are expressed as No. (%).
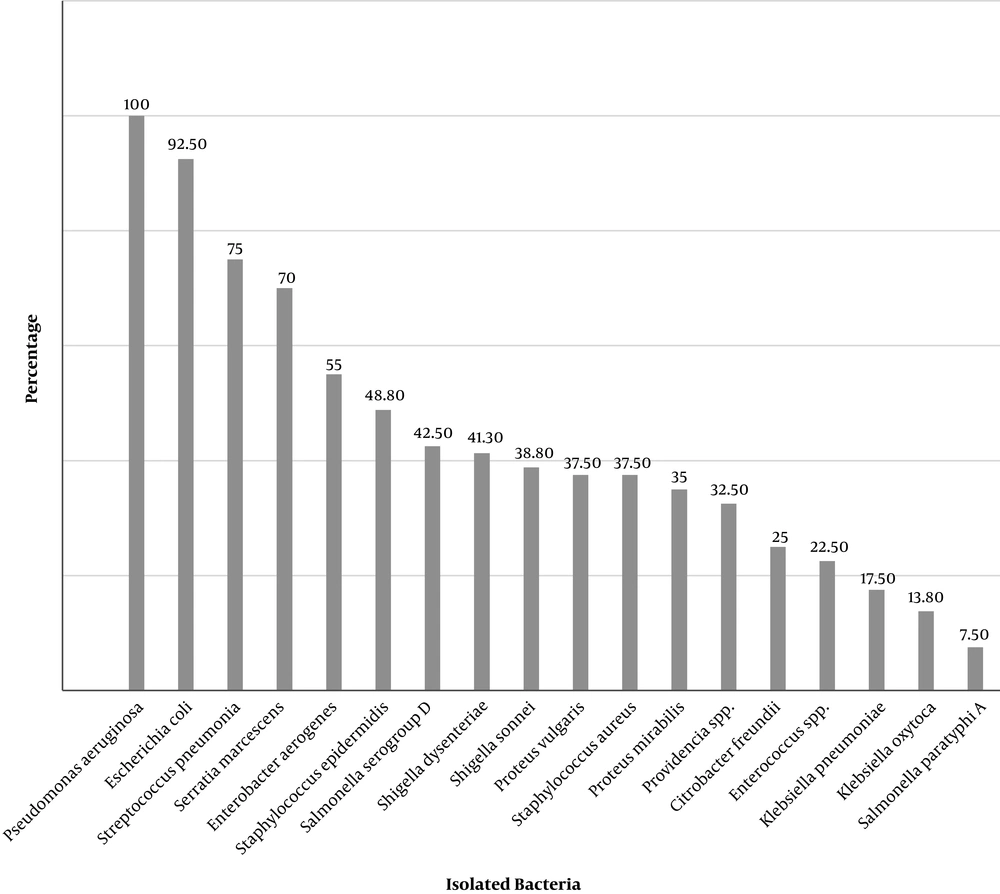
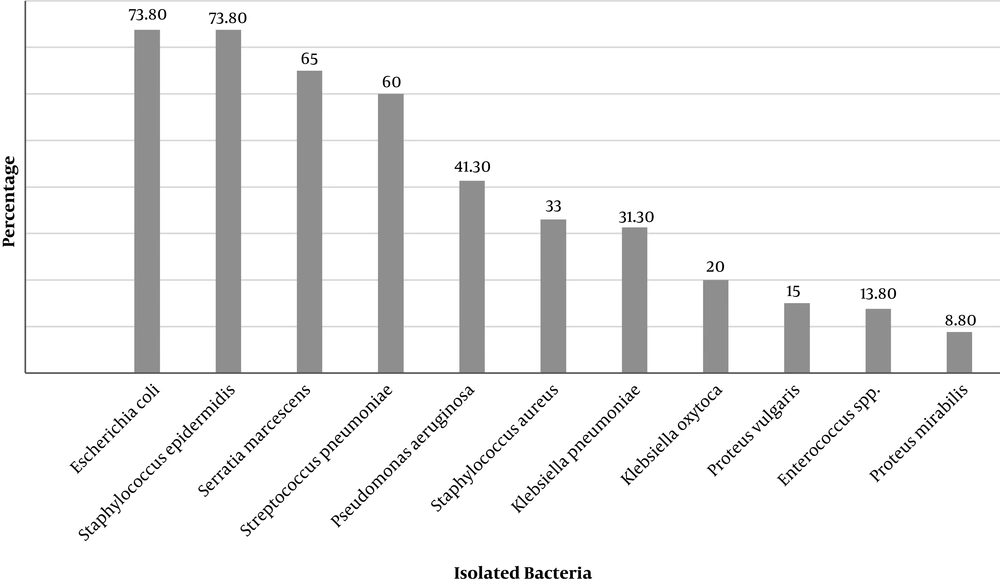
Isolated Gram-negative bacteria from both external surface and the digestive tract of house flies were Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, Proteus mirabilis, Proteus vulgaris, Serratia marcescens, Pseudomonas aeruginosa, Providencia, Enterobacter aerogenes, Shigella sonnei, Salmonella serogroup D, Salmonella paratyphi A, Shigella dysenteriae, and Citrobacter freundii. However, Gram-positive bacteria such as Staphylococcus epidermidis, Streptococcus pneumoniae, Staphylococcus aureus, and Enterococcus were also detected (Figures 4 and 5). In all captures, Staphylococcus epidermidis, Streptococcus pneumoniae, E. coli, and Serratia marcescens were isolated from the external surfaces of the body (Figure 5).
Providencia, Enterobacter aerogenes, Shigella sonnei, Salmonella serogroup D, Salmonella paratyphi A, Shigella dysenteriae, and Citrobacter freundii were only detected from the captured house flies’ digestive tract around the animal husbandry units (Table 1 and Figure 4). In addition, all isolated strains from external parts of these flies were also isolated from their digestive tracts.
Overall, the highest contamination rates of both the external surface and the digestive tracts of house flies were related to the animal husbandry units of the Markazi District of Qom. The animal husbandry units of the other area (e.g., Salafchegan, Khalajestan, and Kahak) were at a later level of importance (Tables 1 and 2).
The most common pathogens isolated from the house flies’ digestive tracts were Pseudomonas aeruginosa (100%) and E. coli (92.5%), and from exterior surfaces of the body were Staphylococcus epidermidis and E. coli (73.8%) (Figure 4).
Based on the location of the captured house flies and their digestive tracts, the most isolated bacteria were Pseudomonas aeruginosa in Markazi District, Pseudomonas aeruginosa, Salmonella serogroup A, Shigella dysenteriae, and Citrobacter freundii in
the Kahak District, Pseudomonas aeruginosa and Serratia marcescens in the Khalajestan District, and Pseudomonas aeruginosa and E. coli and in the Salafchegan District were (Table 1). Based on the captured region and external surfaces, the most isolated bacteria from the Markazi, Kahak, Khalajestan, and Salafchegan districts were Serratia marcescens, Staphylococcus aureus, E. coli, and Staphylococcus epidermidis (Figure 5).
5. Discussion
House flies belong to a species of the insects found throughout the world and because of their great importance in the mechanical transmission of the pathogens, they have been considered as high-risk vectors in many countries (17). Hence, in most studies related to the phonological and biological fields of the important medical flies in Iran, house flies have been reported to exist (18-20). Previously, some studies regarding the transmission of the bacteria and other microorganisms by M. domestica in Iran were published (21, 22), however, our study is the first in the Qom Province, Central Iran. According to previous studies in Iran, E. coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Proteus mirabilis, and Staphylococci aureus have been reported from the external surfaces of the house flies (22, 23). In a similar study conducted in southwest Iran, Kassiri et al. (21), reported some of the isolated bacteria in this present study from house flies at the genus level. The bacterial species that were isolated from house flies in this study were more than the number reported in other studies. Staphylococcus epidermidis, Streptococcus pneumoniae, Klebsiella oxytoca, Enterococcus spp., and Proteus vulgaris are reported for the first time from the surfaces of house flies in Iran. However, some of these species have been reported from other areas around the world (8). The evaluation of the digestive tracts of house flies in this study is the first in Iran. The consideration of the isolated bacteria in the digestive tract of house flies is one of the most controversial issues among researchers (8). The significance of the findings of this study is that some scientists believe that the bacteria in their digestive tracts are more than the external surfaces of their bodies (24). The findings of this study also demonstrate that the isolated bacteria of house flies’ digestive tracts were higher than the external surface of their bodies. Although most of these species have been reported elsewhere in the world, all identified bacteria from the digestive tracks of house flies in this research are reported for the first time in Iran (8). The presence of antimicrobial peptides such as defensin, diptericin and cecropin in the digestive tracks of house flies has been proven (25). It should be noted that considering the presence or absence of pathogenic bacteria in the entire house flies’ digestive tracts (foregut, mid gut, and hindgut) in this study, we cannot correctly declare if bacteria are killed during the house fly’s digestive tracts or not. In terms of epidemiology, this is a very important point. In some studies, the detection of the bacteria from flies has been performed by molecular methods. These assays for detecting bacteria may eclipse the results and interpretations of the tests (8). If the identification method of the flies’ bacterial contamination is solely done based on molecular methods, the presence of the DNA fragments indicates that there are bacteria; however, the differentiation of the live and dead cells is not clear. Nonetheless, in the present study the identification of the bacteria from digestive tracts and external surfaces of house fly’s bodies were done using biochemical profiles that indicated the isolated bacteria were alive and confirmed the gut tract of the house flies can be a source of pathogenic bacteria such as Salmonella, Shigella, etc. Thus, it seems that during the passage of the bacteria through the gut of the flies, a few bacteria can be transmitted via feces to humans and animals (fecal-oral route). In summary, due to the following high-risk factors in the Qom Province, the possibility of mechanical disease transmission and outbreak of communicable disease by flies is increased: (A) a high load of people travel to Qom as a religious city, especially in the summer; (B) most of these pilgrims stay for several days; and (C) this huge population makes places tightly crowded in a high temperature.
5.1. Conclusions
The results of the present study prove that the evaluated insects are potential vectors of the pathogenic strains of the bacterial diseases that can be frequently transmitted to humans and animals. Therefore, controlling the flies is recommended to prevent fly-borne diseases. Finally, the contamination of the house flies cannot be limited to bacteria. Nevertheless, other microorganisms such as parasites, viruses, and fungi may be transmitted by them in the province; thus, further studies are required to detect other pathogens.